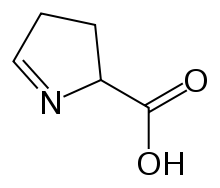
1-Pyrroline-5-carboxylic acid (systematic name 3,4-dihydro-2H-pyrrole-2-carboxylic acid[2]) is a cyclic imino acid. Its conjugate base and anion is 1-pyrroline-5-carboxylate (P5C). In solution, P5C is in spontaneous equilibrium with glutamate-5-semialdhyde (GSA).[3]
| |
| Names | |
|---|---|
| Preferred IUPAC name
3,4-Dihydro-2H-pyrrole-2-carboxylic acid | |
| Other names
1-Pyrroline-5-carboxylic acid
δ-1-Pyrroline-5-carboxylic acid P5C | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| KEGG | |
| MeSH | Delta-1-pyrroline-5-carboxylate |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C5H7NO2 | |
| Molar mass | 113.115 g/mol |
| Acidity (pKa) | 1.82/6.07[1] |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Biochemistry
editThe stereoisomer (S)-1-pyrroline-5-carboxylate (also referred to as L-P5C) is an intermediate metabolite in the biosynthesis and degradation of proline and arginine.[4][5][6]
In prokaryotic proline biosynthesis, GSA is synthesized from γ-glutamyl phosphate by the enzyme γ-glutamyl phosphate reductase. In most eukaryotes, GSA is synthesised from the amino acid glutamate by the bifunctional enzyme 1-pyrroline-5-carboxylate synthase (P5CS). The human P5CS is encoded by the ALDH18A1 gene.[7][8] The enzyme pyrroline-5-carboxylate reductase converts P5C into proline.
In proline degradation, the enzyme proline dehydrogenase produces P5C from proline, and the enzyme 1-pyrroline-5-carboxylate dehydrogenase converts GSA to glutamate. In many prokaryotes, proline dehydrogenase and P5C dehydrogenase form a bifunctional enzyme that prevents the release of P5C during proline degradation.[9]
A reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase (P5CS) and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants proportional to the level of proline.[10] This allows an optimum level of proline to be produced from reduced nitrogen to control osmotic stress.
References
edit- ^ "computed by Chemicalize from ChemAxon".
- ^ PubChem. "3,4-Dihydro-2H-pyrrole-2-carboxylic acid". pubchem.ncbi.nlm.nih.gov. Retrieved 2020-01-23.
- ^ Heacock, Anne M.; Williams, Irene H.; Frank, Leonard H.; Adams, Elijah (1975-04-01). "Δ1-Pyrroline-5-carboxylate and Δ1-pyrroline-3-hydroxy-5-carboxylate: Chromatography on the amino acid analyzer". Analytical Biochemistry. 64 (2): 593–600. doi:10.1016/0003-2697(75)90472-8. ISSN 0003-2697. PMID 236687.
- ^ Bertolo, Robert F.; Burrin, Douglas G. (2008-10-01). "Comparative Aspects of Tissue Glutamine and Proline Metabolism". The Journal of Nutrition. 138 (10): 2032S–2039S. doi:10.1093/jn/138.10.2032S. ISSN 0022-3166. PMID 18806120.
- ^ Qamar, Aarzoo; Mysore, Kirankumar; Senthil-Kumar, Muthappa (2015). "Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens". Frontiers in Plant Science. 6: 503. doi:10.3389/fpls.2015.00503. ISSN 1664-462X. PMC 4491715. PMID 26217357.
- ^ Winter, Gudrun; Todd, Christopher D.; Trovato, Maurizio; Forlani, Giuseppe; Funck, Dietmar (2015). "Physiological implications of arginine metabolism in plants". Frontiers in Plant Science. 6: 534. doi:10.3389/fpls.2015.00534. ISSN 1664-462X. PMC 4520006. PMID 26284079.
- ^ Liu G, Maunoury C, Kamoun P, Aral B (Oct 1996). "Assignment of the human gene encoding the delta 1-pyrroline-5-carboxylate synthetase (P5CS) to 10q24.3 by in situ hybridization". Genomics. 37 (1): 145–6. doi:10.1006/geno.1996.0535. PMID 8921385.
- ^ "Entrez Gene: ALDH18A1 aldehyde dehydrogenase 18 family, member A1".
- ^ Liu, Li-Kai; Becker, Donald F.; Tanner, John J. (2017-10-15). "Structure, function, and mechanism of proline utilization A (PutA)". Archives of Biochemistry and Biophysics. Flavoproteins: Beyond the Classical Paradigms. 632: 142–157. doi:10.1016/j.abb.2017.07.005. ISSN 0003-9861. PMC 5650515. PMID 28712849.
- ^ Verma, D.P.S (December 1996). "Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants". Molecular Genetics. 253 (3): 334–341.