A cancer biomarker refers to a substance or process that is indicative of the presence of cancer in the body. A biomarker may be a molecule secreted by a tumor or a specific response of the body to the presence of cancer. Genetic,[1] epigenetic,[2] proteomic,[3] glycomic,[4] and imaging biomarkers can be used for cancer diagnosis, prognosis, and epidemiology. Ideally, such biomarkers can be assayed in non-invasively collected biofluids like blood or serum.[5]

While numerous challenges exist in translating biomarker research into the clinical space; a number of gene and protein based biomarkers have already been used at some point in patient care; including, AFP (liver cancer), BCR-ABL (chronic myeloid leukemia), BRCA1 / BRCA2 (breast/ovarian cancer), BRAF V600E (melanoma/colorectal cancer), CA-125 (ovarian cancer), CA19.9 (pancreatic cancer), CEA (colorectal cancer), EGFR (Non-small-cell lung carcinoma), HER-2 (Breast Cancer), KIT (gastrointestinal stromal tumor), PSA (prostate specific antigen) (prostate cancer), S100 (melanoma), and many others.[6][7][8][9][10][11][12][13][14][15] Mutant proteins themselves detected by selected reaction monitoring (SRM) have been reported to be the most specific biomarkers for cancers because they can only come from an existing tumor.[16] About 40% of cancers can be cured if detected early through examinations.[17]
Definitions of cancer biomarkers
editOrganizations and publications vary in their definition of biomarker. In many areas of medicine, biomarkers are limited to proteins identifiable or measurable in the blood or urine. However, the term is often used to cover any molecular, biochemical, physiological, or anatomical property that can be quantified or measured.
The National Cancer Institute (NCI), in particular, defines biomarker as a: “A biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease. A biomarker may be used to see how well the body responds to a treatment for a disease or condition. Also called molecular marker and signature molecule."[18]
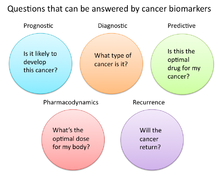
In cancer research and medicine, biomarkers are used in three primary ways:[19]
- To help diagnose conditions, as in the case of identifying early stage cancers (diagnostic)
- To forecast how aggressive a condition is, as in the case of determining a patient's ability to fare in the absence of treatment (prognostic)
- To predict how well a patient will respond to treatment (predictive)
Role of biomarkers in cancer research and medicine
editUses of biomarkers in cancer medicine
editRisk assessment
editCancer biomarkers, particular those associated with genetic mutations or epigenetic alterations, often offer a quantitative way to determine when individuals are predisposed to particular types of cancers. Notable examples of potentially predictive cancer biomarkers include mutations on genes KRAS, p53, EGFR, erbB2 for colorectal, esophageal, liver, and pancreatic cancer; mutations of genes BRCA1 and BRCA2 for breast and ovarian cancer; abnormal methylation of tumor suppressor genes p16, CDKN2B, and p14ARF for brain cancer; hypermethylation of MYOD1, CDH1, and CDH13 for cervical cancer; and hypermethylation of p16, p14, and RB1, for oral cancer.[20]
Diagnosis
editCancer biomarkers can also be useful in establishing a specific diagnosis. This is particularly the case when there is a need to determine whether tumors are of primary or metastatic origin. To make this distinction, researchers can screen the chromosomal alterations found on cells located in the primary tumor site against those found in the secondary site. If the alterations match, the secondary tumor can be identified as metastatic; whereas if the alterations differ, the secondary tumor can be identified as a distinct primary tumor.[21] For example, people with tumors have high levels of circulating tumor DNA (ctDNA) due to tumor cells that have gone through apoptosis.[22] This tumor marker can be detected in the blood, saliva, or urine.[17] The possibility of identifying an effective biomarker for early cancer diagnosis has recently been questioned, in light of the high molecular heterogeneity of tumors observed by next-generation sequencing studies.[23]
Prognosis and treatment predictions
editAnother use of biomarkers in cancer medicine is for disease prognosis, which take place after an individual has been diagnosed with cancer. Here biomarkers can be useful in determining the aggressiveness of an identified cancer as well as its likelihood of responding to a given treatment. In part, this is because tumors exhibiting particular biomarkers may be responsive to treatments tied to that biomarker's expression or presence. Examples of such prognostic biomarkers include elevated levels of metallopeptidase inhibitor 1 (TIMP1), a marker associated with more aggressive forms of multiple myeloma,[24] elevated estrogen receptor (ER) and/or progesterone receptor (PR) expression, markers associated with better overall survival in patients with breast cancer;[25][26] HER2/neu gene amplification, a marker indicating a breast cancer will likely respond to trastuzumab treatment;[27][28] a mutation in exon 11 of the proto-oncogene c-KIT, a marker indicating a gastrointestinal stromal tumor (GIST) will likely respond to imatinib treatment;[29][30] and mutations in the tyrosine kinase domain of EGFR1, a marker indicating a patient's non-small-cell lung carcinoma (NSCLC) will likely respond to gefitinib or erlotinib treatment.[31][32]
Pharmacodynamics and pharmacokinetics
editCancer biomarkers can also be used to determine the most effective treatment regime for a particular person's cancer.[33] Because of differences in each person's genetic makeup, some people metabolize or change the chemical structure of drugs differently. In some cases, decreased metabolism of certain drugs can create dangerous conditions in which high levels of the drug accumulate in the body. As such, drug dosing decisions in particular cancer treatments can benefit from screening for such biomarkers. An example is the gene encoding the enzyme thiopurine methyl-transferase (TPMPT).[34] Individuals with mutations in the TPMT gene are unable to metabolize large amounts of the leukemia drug, mercaptopurine, which potentially causes a fatal drop in white blood count for such patients. Patients with TPMT mutations are thus recommended to be given a lower dose of mercaptopurine for safety considerations.[35]
Monitoring treatment response
editCancer biomarkers have also shown utility in monitoring how well a treatment is working over time. Much research is going into this particular area, since successful biomarkers have the potential of providing significant cost reduction in patient care, as the current image-based tests such as CT and MRI for monitoring tumor status are highly costly.[36]
One notable biomarker garnering significant attention is the protein biomarker S100-beta in monitoring the response of malignant melanoma. In such melanomas, melanocytes, the cells that make pigment in our skin, produce the protein S100-beta in high concentrations dependent on the number of cancer cells. Response to treatment is thus associated with reduced levels of S100-beta in the blood of such individuals.[37][38]
Similarly, additional laboratory research has shown that tumor cells undergoing apoptosis can release cellular components such as cytochrome c, nucleosomes, cleaved cytokeratin-18, and E-cadherin. Studies have found that these macromolecules and others can be found in circulation during cancer therapy, providing a potential source of clinical metrics for monitoring treatment.[36]
Recurrence
editCancer biomarkers can also offer value in predicting or monitoring cancer recurrence. The Oncotype DX® breast cancer assay is one such test used to predict the likelihood of breast cancer recurrence. This test is intended for women with early-stage (Stage I or II), node-negative, estrogen receptor-positive (ER+) invasive breast cancer who will be treated with hormone therapy. Oncotype DX looks at a panel of 21 genes in cells taken during tumor biopsy. The results of the test are given in the form of a recurrence score that indicates likelihood of recurrence at 10 years.[39][40]
Uses of biomarkers in cancer research
editDeveloping drug targets
editIn addition to their use in cancer medicine, biomarkers are often used throughout the cancer drug discovery process. For instance, in the 1960s, researchers discovered the majority of patients with chronic myelogenous leukemia possessed a particular genetic abnormality on chromosomes 9 and 22 dubbed the Philadelphia chromosome. When these two chromosomes combine they create a cancer-causing gene known as BCR-ABL. In such patients, this gene acts as the principle initial point in all of the physiological manifestations of the leukemia. For many years, the BCR-ABL was simply used as a biomarker to stratify a certain subtype of leukemia. However, drug developers were eventually able to develop imatinib, a powerful drug that effectively inhibited this protein and significantly decreased production of cells containing the Philadelphia chromosome.[41][42]
Surrogate endpoints
editAnother promising area of biomarker application is in the area of surrogate endpoints. In this application, biomarkers act as stand-ins for the effects of a drug on cancer progression and survival. Ideally, the use of validated biomarkers would prevent patients from having to undergo tumor biopsies and lengthy clinical trials to determine if a new drug worked. In the current standard of care, the metric for determining a drug's effectiveness is to check if it has decreased cancer progression in humans and ultimately whether it prolongs survival. However, successful biomarker surrogates could save substantial time, effort, and money if failing drugs could be eliminated from the development pipeline before being brought to clinical trials.
Some ideal characteristics of surrogate endpoint biomarkers include:[43][44]
- Biomarker should be involved in process that causes the cancer
- Changes in biomarker should correlate with changes in the disease
- Levels of biomarkers should be high enough that they can be measured easily and reliably
- Levels or presence of biomarker should readily distinguish between normal, cancerous, and precancerous tissue
- Effective treatment of the cancer should change the level of the biomarker
- Level of the biomarker should not change spontaneously or in response to other factors not related to the successful treatment of the cancer
Two areas in particular that are receiving attention as surrogate markers include circulating tumor cells (CTCs)[45][46] and circulating miRNAs.[47][48] Both these markers are associated with the number of tumor cells present in the blood, and as such, are hoped to provide a surrogate for tumor progression and metastasis. However, significant barriers to their adoption include the difficulty of enriching, identifying, and measuring CTC and miRNA levels in blood. New technologies and research are likely necessary for their translation into clinical care.[49][50][51]
Types of cancer biomarkers
editMolecular cancer biomarkers
edit| Tumor type | Biomarker |
|---|---|
| Breast | ER/PR (estrogen receptor/progesteron receptor)[52][53] |
| HER-2/neu[52][53] | |
| Colorectal | EGFR[52][53] |
| KRAS[52][54] | |
| UGT1A1[52][54] | |
| Gastric | HER-2/neu [52] |
| GIST | c-KIT[52][55] |
| Leukemia/lymphoma | CD20[52][56] |
| CD30[52][57] | |
| FIP1L1-PDGFRalpha[52][58] | |
| PDGFR[52][59] | |
| Philadelphia chromosome (BCR/ABL) [52][60][61] | |
| PML/RAR-alpha[52][62] | |
| TPMT[52][63] | |
| UGT1A1 [52][64] | |
| Lung | EML4/ALK[52][65][66] |
| EGFR [52][53] | |
| KRAS [52][53] | |
| Melanoma | BRAF[52][66] |
| Pancreas | Elevated levels of leucine, isoleucine and valine[67] |
| Ovaries | CA-125[68] |
Other examples of biomarkers:
Cancer biomarkers without specificity
editNot all cancer biomarkers have to be specific to types of cancer. Some biomarkers found in the circulatory system can be used to determine an abnormal growth of cells present in the body. All these types of biomarkers can be identified through diagnostic blood tests, which is one of the main reasons to get regularly health tested. By getting regularly tested, many health issues such as cancer can be discovered at an early stage, preventing many deaths.
The neutrophil-to-lymphocyte ratio has been shown to be a non-specific determinant for many cancers. This ratio focuses on the activity of two components of the immune system that are involved in inflammatory response which is shown to be higher in presence of malignant tumors.[71] Additionally, basic fibroblast growth factor (bFGF) is a protein that is involved in the proliferation of cells. Unfortunately, it has been shown that in the presence of tumors it is highly active which has led to the conclusion that it may help malignant cells reproduce at faster rates.[72] Research has shown that anti-bFGF antibodies can be used to help treat tumors from many origins.[72] Moreover, insulin-like growth factor (IGF-R) is involved in cell proliferation and growth. It has is possible that it is involved in inhibiting apoptosis, programmed cell death due to some defect.[73] Due to this, the levels of IGF-R can be increased when cancer such as breast, prostate, lung, and colorectum is present.[74]
| Biomarker | Description | Biosensor used |
|---|---|---|
| NLR (neutrophil-to-lymphocyte ratio) | Elevates with inflammation caused by cancer[75] | No |
| Basic Fibroblast Growth Factor (bFGF) | This level increases when a tumor is present, helps with the fast reproduction of tumor cells[76] | Electrochemical[77] |
| Insulin-like Growth Factor (IGF-R) | High activity in cancer cells, help reproduction[78] | Electrochemical Impedance Spectroscopy Sensor[79] |
See also
editReferences
edit- ^ Calzone KA (May 2012). "Genetic biomarkers of cancer risk". Seminars in Oncology Nursing. 28 (2): 122–128. doi:10.1016/j.soncn.2012.03.007. PMC 10433658. PMID 22542320.
- ^ Herceg Z, Hainaut P (June 2007). "Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis". Molecular Oncology. 1 (1): 26–41. doi:10.1016/j.molonc.2007.01.004. PMC 5543860. PMID 19383285.
- ^ Li D, Chan DW (April 2014). "Proteomic cancer biomarkers from discovery to approval: it's worth the effort". Expert Review of Proteomics. 11 (2): 135–136. doi:10.1586/14789450.2014.897614. PMC 4079106. PMID 24646122.
- ^ Aizpurua-Olaizola O, Toraño JS, Falcon-Perez JM, Williams C, Reichardt N, Boons GJ (2018). "Mass spectrometry for glycan biomarker discovery". TrAC Trends in Analytical Chemistry. 100: 7–14. doi:10.1016/j.trac.2017.12.015.
- ^ Mishra A, Verma M (March 2010). "Cancer biomarkers: are we ready for the prime time?". Cancers. 2 (1): 190–208. doi:10.3390/cancers2010190. PMC 3827599. PMID 24281040.
- ^ Rhea J, Molinaro RJ (March 2011). "Cancer Biomarkers: Surviving the journey from bench to bedside". Medical Laboratory Observer. Archived from the original on 14 October 2013. Retrieved 26 April 2013.
- ^ Behne T, Copur MS (1 January 2012). "Biomarkers for hepatocellular carcinoma". International Journal of Hepatology. 2012: 859076. doi:10.1155/2012/859076. PMC 3357951. PMID 22655201.
- ^ Musolino A, Bella MA, Bortesi B, Michiara M, Naldi N, Zanelli P, et al. (June 2007). "BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: a population-based study". Breast. 16 (3): 280–292. doi:10.1016/j.breast.2006.12.003. hdl:11381/1629553. PMID 17257844.
- ^ Dienstmann R, Tabernero J (March 2011). "BRAF as a target for cancer therapy". Anti-Cancer Agents in Medicinal Chemistry. 11 (3): 285–295. doi:10.2174/187152011795347469. PMID 21426297.
- ^ Lamparella N, Barochia A, Almokadem S (2013). "Impact of Genetic Markers on Treatment of Non-small Cell Lung Cancer". Impact of Genetic Targets on Cancer Therapy. Advances in Experimental Medicine and Biology. Vol. 779. pp. 145–164. doi:10.1007/978-1-4614-6176-0_6. ISBN 978-1-4614-6175-3. PMID 23288638.
- ^ Orphanos G, Kountourakis P (2012). "Targeting the HER2 receptor in metastatic breast cancer". Hematology/Oncology and Stem Cell Therapy. 5 (3): 127–137. doi:10.5144/1658-3876.2012.127. PMID 23095788.
- ^ Deprimo SE, Huang X, Blackstein ME, Garrett CR, Harmon CS, Schöffski P, et al. (September 2009). "Circulating levels of soluble KIT serve as a biomarker for clinical outcome in gastrointestinal stromal tumor patients receiving sunitinib following imatinib failure". Clinical Cancer Research. 15 (18): 5869–5877. doi:10.1158/1078-0432.CCR-08-2480. PMC 3500590. PMID 19737953.
- ^ Bantis A, Grammaticos P (Sep–Dec 2012). "Prostatic specific antigen and bone scan in the diagnosis and follow-up of prostate cancer. Can diagnostic significance of PSA be increased?". Hellenic Journal of Nuclear Medicine. 15 (3): 241–246. PMID 23227460.
- ^ Kruijff S, Hoekstra HJ (April 2012). "The current status of S-100B as a biomarker in melanoma". European Journal of Surgical Oncology. 38 (4): 281–285. doi:10.1016/j.ejso.2011.12.005. PMID 22240030.
- ^ Ludwig JA, Weinstein JN (November 2005). "Biomarkers in cancer staging, prognosis and treatment selection". Nature Reviews. Cancer. 5 (11): 845–856. doi:10.1038/nrc1739. PMID 16239904. S2CID 25540232.
- ^ Wang Q, Chaerkady R, Wu J, Hwang HJ, Papadopoulos N, Kopelovich L, et al. (February 2011). "Mutant proteins as cancer-specific biomarkers". Proceedings of the National Academy of Sciences of the United States of America. 108 (6): 2444–2449. Bibcode:2011PNAS..108.2444W. doi:10.1073/pnas.1019203108. PMC 3038743. PMID 21248225.
- ^ a b c Li X, Ye M, Zhang W, Tan D, Jaffrezic-Renault N, Yang X, Guo Z (February 2019). "Liquid biopsy of circulating tumor DNA and biosensor applications". Biosensors & Bioelectronics. 126: 596–607. doi:10.1016/j.bios.2018.11.037. PMID 30502682. S2CID 56479882.
- ^ "biomarker". NCI Dictionary of Cancer Terms. National Cancer Institute. 2011-02-02.
- ^ "Biomarkers in Cancer: An Introductory Guide for Advocates" (PDF). Research Advocacy Network. 2010. Archived from the original (PDF) on 2013-10-29. Retrieved 26 April 2013.
- ^ Verma M, Manne U (October 2006). "Genetic and epigenetic biomarkers in cancer diagnosis and identifying high risk populations". Critical Reviews in Oncology/Hematology. 60 (1): 9–18. doi:10.1016/j.critrevonc.2006.04.002. PMID 16829121.
- ^ Leong PP, Rezai B, Koch WM, Reed A, Eisele D, Lee DJ, et al. (July 1998). "Distinguishing second primary tumors from lung metastases in patients with head and neck squamous cell carcinoma". Journal of the National Cancer Institute. 90 (13): 972–977. doi:10.1093/jnci/90.13.972. PMID 9665144.
- ^ Lapin M, Oltedal S, Tjensvoll K, Buhl T, Smaaland R, Garresori H, et al. (November 2018). "Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer". Journal of Translational Medicine. 16 (1): 300. doi:10.1186/s12967-018-1677-2. PMC 6218961. PMID 30400802.
- ^ Dragani TA, Matarese V, Colombo F (April 2020). "Biomarkers for Early Cancer Diagnosis: Prospects for Success through the Lens of Tumor Genetics". BioEssays. 42 (4): e1900122. doi:10.1002/bies.201900122. PMID 32128843. S2CID 212406467.
- ^ Terpos E, Dimopoulos MA, Shrivastava V, Leitzel K, Christoulas D, Migkou M, et al. (March 2010). "High levels of serum TIMP-1 correlate with advanced disease and predict for poor survival in patients with multiple myeloma treated with novel agents". Leukemia Research. 34 (3): 399–402. doi:10.1016/j.leukres.2009.08.035. PMID 19781774.
- ^ Kuukasjärvi T, Kononen J, Helin H, Holli K, Isola J (September 1996). "Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy". Journal of Clinical Oncology. 14 (9): 2584–2589. doi:10.1200/jco.1996.14.9.2584. PMID 8823339.
- ^ Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. (November 2007). "American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer". Journal of Clinical Oncology. 25 (33): 5287–5312. doi:10.1200/JCO.2007.14.2364. PMID 17954709.
- ^ Kröger N, Milde-Langosch K, Riethdorf S, Schmoor C, Schumacher M, Zander AR, Löning T (January 2006). "Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients". Clinical Cancer Research. 12 (1): 159–168. doi:10.1158/1078-0432.CCR-05-1340. PMID 16397038.
- ^ Vrbic S, Pejcic I, Filipovic S, Kocic B, Vrbic M (Jan–Mar 2013). "Current and future anti-HER2 therapy in breast cancer". Journal of B.U.On. 18 (1): 4–16. PMID 23613383.
- ^ Yoo C, Ryu MH, Ryoo BY, Beck MY, Kang YK (October 2013). "Efficacy, safety, and pharmacokinetics of imatinib dose escalation to 800 mg/day in patients with advanced gastrointestinal stromal tumors". Investigational New Drugs. 31 (5): 1367–1374. doi:10.1007/s10637-013-9961-8. PMID 23591629. S2CID 29477955.
- ^ Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, et al. (October 2006). "Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial". Lancet. 368 (9544): 1329–1338. doi:10.1016/S0140-6736(06)69446-4. PMID 17046465. S2CID 25931515.
- ^ Herbst RS, Prager D, Hermann R, Fehrenbacher L, Johnson BE, Sandler A, et al. (September 2005). "TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer". Journal of Clinical Oncology. 23 (25): 5892–5899. doi:10.1200/JCO.2005.02.840. PMID 16043829.
- ^ Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. (May 2004). "Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib" (PDF). The New England Journal of Medicine. 350 (21): 2129–2139. doi:10.1056/NEJMoa040938. PMID 15118073.
- ^ Sawyers CL (April 2008). "The cancer biomarker problem". Nature. 452 (7187): 548–552. Bibcode:2008Natur.452..548S. doi:10.1038/nature06913. PMID 18385728. S2CID 205213083.
- ^ Karas-Kuzelicki N, Mlinaric-Rascan I (August 2009). "Individualization of thiopurine therapy: thiopurine S-methyltransferase and beyond". Pharmacogenomics. 10 (8): 1309–1322. doi:10.2217/pgs.09.78. PMID 19663675.
- ^ Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, et al. (December 1999). "Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus". Journal of the National Cancer Institute. 91 (23): 2001–2008. doi:10.1093/jnci/91.23.2001. PMID 10580024.
- ^ a b Schneider JE, Sidhu MK, Doucet C, Kiss N, Ohsfeldt RL, Chalfin D (November 2012). "Economics of cancer biomarkers". Personalized Medicine. 9 (8): 829–837. doi:10.2217/pme.12.87. PMID 29776231.
- ^ Henze G, Dummer R, Joller-Jemelka HI, Böni R, Burg G (1997). "Serum S100--a marker for disease monitoring in metastatic melanoma". Dermatology. 194 (3): 208–212. doi:10.1159/000246103. PMID 9187834.
- ^ Harpio R, Einarsson R (July 2004). "S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma". Clinical Biochemistry. 37 (7): 512–518. doi:10.1016/j.clinbiochem.2004.05.012. PMID 15234232.
- ^ Lamond NW, Skedgel C, Younis T (April 2013). "Is the 21-gene recurrence score a cost-effective assay in endocrine-sensitive node-negative breast cancer?". Expert Review of Pharmacoeconomics & Outcomes Research. 13 (2): 243–250. doi:10.1586/erp.13.4. PMID 23570435. S2CID 33661439.
- ^ Biroschak JR, Schwartz GF, Palazzo JP, Toll AD, Brill KL, Jaslow RJ, Lee SY (May 2013). "Impact of Oncotype DX on treatment decisions in ER-positive, node-negative breast cancer with histologic correlation". The Breast Journal. 19 (3): 269–275. doi:10.1111/tbj.12099. PMID 23614365. S2CID 30895945.
- ^ Moen MD, McKeage K, Plosker GL, Siddiqui MA (2007). "Imatinib: a review of its use in chronic myeloid leukaemia". Drugs. 67 (2): 299–320. doi:10.2165/00003495-200767020-00010. PMID 17284091.
- ^ Lemonick M, Park A (May 28, 2001). "New Hope for Cancer". Time. Archived from the original on October 15, 2007. Retrieved 26 April 2013.
- ^ Price C, McDonnell D (February 1991). "Effects of niobium filtration and constant potential on the sensitometric responses of dental radiographic films". Dento Maxillo Facial Radiology. 20 (1): 11–16. doi:10.1259/dmfr.20.1.1884846 (inactive 2024-11-28). PMID 1884846.
{{cite journal}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Cohen V, Khuri FR (2003). "Progress in lung cancer chemoprevention". Cancer Control. 10 (4): 315–324. doi:10.1177/107327480301000406. PMID 12915810.
- ^ Lu CY, Tsai HL, Uen YH, Hu HM, Chen CW, Cheng TL, et al. (March 2013). "Circulating tumor cells as a surrogate marker for determining clinical outcome to mFOLFOX chemotherapy in patients with stage III colon cancer". British Journal of Cancer. 108 (4): 791–797. doi:10.1038/bjc.2012.595. PMC 3590657. PMID 23422758.
- ^ Balic M, Williams A, Lin H, Datar R, Cote RJ (2013). "Circulating tumor cells: from bench to bedside". Annual Review of Medicine. 64: 31–44. doi:10.1146/annurev-med-050311-163404. PMC 3809995. PMID 23092385.
- ^ Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, et al. (November 2012). "Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer". Clinical Cancer Research. 18 (21): 5972–5982. doi:10.1158/1078-0432.CCR-12-1407. PMID 22952344.
- ^ Redova M, Sana J, Slaby O (March 2013). "Circulating miRNAs as new blood-based biomarkers for solid cancers". Future Oncology. 9 (3): 387–402. doi:10.2217/fon.12.192. PMID 23469974.
- ^ Joosse SA, Pantel K (January 2013). "Biologic challenges in the detection of circulating tumor cells". Cancer Research. 73 (1): 8–11. doi:10.1158/0008-5472.CAN-12-3422. PMID 23271724.
- ^ Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, et al. (2013). "Isolation and retrieval of circulating tumor cells using centrifugal forces". Scientific Reports. 3: 1259. Bibcode:2013NatSR...3.1259H. doi:10.1038/srep01259. PMC 3569917. PMID 23405273.
- ^ Dhondt B, De Bleser E, Claeys T, Buelens S, Lumen N, Vandesompele J, et al. (December 2019). "Discovery and validation of a serum microRNA signature to characterize oligo- and polymetastatic prostate cancer: not ready for prime time". World Journal of Urology. 37 (12): 2557–2564. doi:10.1007/s00345-018-2609-8. hdl:1854/LU-8586484. PMID 30578441. S2CID 58594673.
- ^ a b c d e f g h i j k l m n o p q r s "Table of Pharmacogenomic Biomarkers in Drug Labels". U.S. Food and Drug Administration.
- ^ a b c d e "Tumor Markers Fact Sheet" (PDF). American Cancer Society.
- ^ a b Heinz-Josef Lenz (2012-09-18). Biomarkers in Oncology: Prediction and Prognosis. Springer Science & Business Media. p. 263. ISBN 978-1-4419-9754-8.
- ^ Gonzalez RS, Carlson G, Page AJ, Cohen C (July 2011). "Gastrointestinal stromal tumor markers in cutaneous melanomas: relationship to prognostic factors and outcome". American Journal of Clinical Pathology. 136 (1): 74–80. doi:10.1309/AJCP9KHD7DCHWLMO. PMID 21685034.
- ^ Tam CS, Otero-Palacios J, Abruzzo LV, Jorgensen JL, Ferrajoli A, Wierda WG, et al. (April 2008). "Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in-situ hybridization in 510 patients". British Journal of Haematology. 141 (1): 36–40. doi:10.1111/j.1365-2141.2008.07012.x. PMID 18324964.
- ^ Zhang M, Yao Z, Patel H, Garmestani K, Zhang Z, Talanov VS, et al. (May 2007). "Effective therapy of murine models of human leukemia and lymphoma with radiolabeled anti-CD30 antibody, HeFi-1". Proceedings of the National Academy of Sciences of the United States of America. 104 (20): 8444–8448. Bibcode:2007PNAS..104.8444Z. doi:10.1073/pnas.0702496104. PMC 1895969. PMID 17488826.
- ^ Yamada Y, Sanchez-Aguilera A, Brandt EB, McBride M, Al-Moamen NJ, Finkelman FD, et al. (September 2008). "FIP1L1/PDGFRalpha synergizes with SCF to induce systemic mastocytosis in a murine model of chronic eosinophilic leukemia/hypereosinophilic syndrome". Blood. 112 (6): 2500–2507. doi:10.1182/blood-2007-11-126268. PMID 18539901.
- ^ Nimer SD (May 2008). "Myelodysplastic syndromes". Blood. 111 (10): 4841–4851. doi:10.1182/blood-2007-08-078139. PMID 18467609. S2CID 6802096.
- ^ Ottmann O, Dombret H, Martinelli G, Simonsson B, Guilhot F, Larson RA, et al. (October 2007). "Dasatinib induces rapid hematologic and cytogenetic responses in adult patients with Philadelphia chromosome positive acute lymphoblastic leukemia with resistance or intolerance to imatinib: interim results of a phase 2 study". Blood. 110 (7): 2309–2315. doi:10.1182/blood-2007-02-073528. PMID 17496201.
- ^ Boulos N, Mulder HL, Calabrese CR, Morrison JB, Rehg JE, Relling MV, et al. (March 2011). "Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia". Blood. 117 (13): 3585–3595. doi:10.1182/blood-2010-08-301267. PMC 3072880. PMID 21263154.
- ^ O'Connell PA, Madureira PA, Berman JN, Liwski RS, Waisman DM (April 2011). "Regulation of S100A10 by the PML-RAR-α oncoprotein". Blood. 117 (15): 4095–4105. doi:10.1182/blood-2010-07-298851. PMID 21310922.
- ^ Duffy MJ, Crown J (November 2008). "A personalized approach to cancer treatment: how biomarkers can help". Clinical Chemistry. 54 (11): 1770–1779. doi:10.1373/clinchem.2008.110056. PMID 18801934.
- ^ Ribrag V, Koscielny S, Casasnovas O, Cazeneuve C, Brice P, Morschhauser F, et al. (April 2009). "Pharmacogenetic study in Hodgkin lymphomas reveals the impact of UGT1A1 polymorphisms on patient prognosis". Blood. 113 (14): 3307–3313. doi:10.1182/blood-2008-03-148874. PMID 18768784.
- ^ Li Y, Ye X, Liu J, Zha J, Pei L (January 2011). "Evaluation of EML4-ALK fusion proteins in non-small cell lung cancer using small molecule inhibitors". Neoplasia. 13 (1): 1–11. doi:10.1593/neo.101120. PMC 3022423. PMID 21245935.
- ^ a b Pao W, Girard N (February 2011). "New driver mutations in non-small-cell lung cancer". The Lancet. Oncology. 12 (2): 175–180. doi:10.1016/S1470-2045(10)70087-5. PMID 21277552.
- ^ Hewes A (October 2, 2014). "Promising Method for Detecting Pancreatic Cancer Years Before Traditional Diagnosis". Singularity HUB. Retrieved 2016-04-22.
- ^ Gupta D, Lis CG (October 2009). "Role of CA125 in predicting ovarian cancer survival - a review of the epidemiological literature". Journal of Ovarian Research. 2 (1): 13. doi:10.1186/1757-2215-2-13. PMC 2764643. PMID 19818123.
- ^ Bartels CL, Tsongalis GJ (April 2009). "MicroRNAs: novel biomarkers for human cancer". Clinical Chemistry. 55 (4): 623–631. doi:10.1373/clinchem.2008.112805. PMID 19246618.
- ^ Paulson KG, Lewis CW, Redman MW, Simonson WT, Lisberg A, Ritter D, et al. (April 2017). "Viral oncoprotein antibodies as a marker for recurrence of Merkel cell carcinoma: A prospective validation study". Cancer. 123 (8): 1464–1474. doi:10.1002/cncr.30475. PMC 5384867. PMID 27925665.
- ^ Proctor MJ, McMillan DC, Morrison DS, Fletcher CD, Horgan PG, Clarke SJ (August 2012). "A derived neutrophil to lymphocyte ratio predicts survival in patients with cancer". British Journal of Cancer. 107 (4): 695–699. doi:10.1038/bjc.2012.292. PMC 3419948. PMID 22828611.
- ^ a b Liu M, Xing LQ (August 2017). "Basic fibroblast growth factor as a potential biomarker for diagnosing malignant tumor metastasis in women". Oncology Letters. 14 (2): 1561–1567. doi:10.3892/ol.2017.6335. PMC 5529833. PMID 28789380.
- ^ Fürstenberger G, Senn HJ (May 2002). "Insulin-like growth factors and cancer". The Lancet. Oncology. 3 (5): 298–302. doi:10.1016/s1470-2045(02)00731-3. PMID 12067807.
- ^ Yu H, Rohan T (September 2000). "Role of the insulin-like growth factor family in cancer development and progression". Journal of the National Cancer Institute. 92 (18): 1472–1489. doi:10.1093/jnci/92.18.1472. PMID 10995803.
- ^ Vano YA, Oudard S, By MA, Têtu P, Thibault C, Aboudagga H, et al. (2018-04-06). "Optimal cut-off for neutrophil-to-lymphocyte ratio: Fact or Fantasy? A prospective cohort study in metastatic cancer patients". PLOS ONE. 13 (4): e0195042. Bibcode:2018PLoSO..1395042V. doi:10.1371/journal.pone.0195042. PMC 5889159. PMID 29624591.
- ^ Liu M, Xing LQ (August 2017). "Basic fibroblast growth factor as a potential biomarker for diagnosing malignant tumor metastasis in women". Oncology Letters. 14 (2): 1561–1567. doi:10.3892/ol.2017.6335. PMC 5529833. PMID 28789380.
- ^ Torrente-Rodríguez RM, Ruiz-Valdepeñas Montiel V, Campuzano S, Pedrero M, Farchado M, Vargas E, et al. (2017-04-04). "Electrochemical sensor for rapid determination of fibroblast growth factor receptor 4 in raw cancer cell lysates". PLOS ONE. 12 (4): e0175056. Bibcode:2017PLoSO..1275056T. doi:10.1371/journal.pone.0175056. PMC 5380347. PMID 28376106.
- ^ Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, et al. (March 2015). "Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance". Genes & Diseases. 2 (1): 13–25. doi:10.1016/j.gendis.2014.10.004. PMC 4431759. PMID 25984556.
- ^ Rezaei, Behzad; Majidi, Najmeh; Rahmani, Hamidreza; Khayamian, Taghi (2011). "Electrochemical impedimetric immunosensor for insulin like growth factor-1 using specific monoclonal antibody-nanogold modified electrode". Biosensors and Bioelectronics. 26 (5). Elsevier BV: 2130–2134. doi:10.1016/j.bios.2010.09.020. ISSN 0956-5663. PMID 20933383.