Sperm microtubule associated protein 1 is a protein which in humans is encoded by the SPMAP1 gene. The protein is derived from Homo sapiens chromosome 17.[5] The SPMAP1 gene consists of a 6,302 base sequence. Its mRNA has three exons and no alternative splice sites. The protein has 154 amino acids, with no abnormal amino acid levels.[6] SPMAP1 has a domain of unknown function (DUF4542) and is 17.6kDa in weight.[7][8] SPMAP1 does not belong to any other families nor does it have any isoforms.[9] The protein has orthologs with high percent similarity in mammals and reptiles. The protein has additional distantly related orthologs across the metazoan kingdom, culminating with the sponge family.[10]
| SPMAP1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | SPMAP1, chromosome 17 open reading frame 98, sperm microtubule associated protein 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1919465; HomoloGene: 19140; GeneCards: SPMAP1; OMA:SPMAP1 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Like most proteins, SPMAP1 is known to be highly expressed in the testes.[11] The protein has also been known to have elevated levels in cancer.[11] The protein has been shown to be expressed in proximity to or within intermediate filaments and the nucleolus.[11] Additionally, SPMAP1 has transcription factors which are also active in hematopoietic stem cells, the immune system, and the cardiovascular system, among others.[12] The gene is over-expressed in many cancer types, including kidney renal clear cell carcinoma and lung squamous cell carcinoma.[13] Motif and transcription factor analysis points towards SPMAP1 playing a role in proliferation, specially in immune cell proliferation.
Gene
editBackground
editThe SPMAP1 gene consists of 6,303 bases. It has three exons and two large introns. The gene has no alternative splice sites.[14] The 5' UTR sequence of SPMAP1 is highly conserved in primates. No non-mammalian 5' UTR matches were able to be determined.[15][16] SPMAP1 has 11 Alu repeats.[17]
Enhancers
editGeneCards determined that SPMAP1 has five enhancer sequences. The role of the sequences may provide insight into the function of SPMAP1. Four of the five enhancers are active in the thymus. All five enhancers are active in the H1 hESC. Additionally, all five enhancers are active in iPS DF 19.11 derived from foreskin fibroblasts.[18]
Transcription factors
editThe SPMAP1 promoter has many transcription factors binding sites.[19] SPMAP1's transcription factors are commonly found in hematopoietic cells, connective tissue, cardiovascular tissue, and the immune system. The presence of Krueppel Like Transcription Factors suggests a role for SPMAP1 in proliferation or apoptosis. The presence of SMAD indicates an involvement in the TGF-β pathway, while the presence of Myc related transcription factors indicates a potential proliferation function of the protein. Additionally, other SPMAP1 transcription factors, like RBPJ-Kappa are involved in proliferation and signalling.
Variants
editNumerous SNPs were found in the 5' UTR, 3' UTR, and coding region of SPMAP1.[20] Few SNPs were found in highly conserved regions. In all, four SNPs were found in the highly conserved amino acids. One SNP was found in the start codon sequence. Of these five, three had a SNP on the third position of the codon. Due to the wobble hypothesis, three of the five SNPs would have no effect on the overall protein structure.
mRNA
editSPMAP1 does not have any miRNA binding sites.[21] Its mRNA has low abundance (0.44%).[22] The mRNA sequence has three hexaloops, none of which are significant.[23]
Protein
editPrimary structure
editSPMAP1 is a 17.6kDa protein.[8] Distant orthologs are 5 to 6 kDa larger, but some of the discrepancies come from an added NLS sequence, which Homo sapiens does not have There are no positive or negative charge clusters. There are no transmembrane components. The isoelectric point is 9.80 / 17564.67 pI/Mw.[24] SPMAP1 is hydrophobic and soluble.
Secondary and tertiary structure
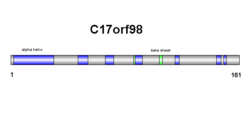
editSecondary structure of SPMAP1 consists of both beta sheets and alpha helices (see diagram on right). Results are confirmed in the tertiary structure, however, alpha helix and beta sheet numbers differ slightly (see diagram on right).
Motifs and binding sites
editThere are no N-terminal signal peptides. Cleavage motifs were not found. There are no ER membrane retention signals, nor peroxisomal targeting signal. SKL2 is not present, thus a secondary peroxisome signal is not present. There are no vacuolar targeting signals. There are no RNA binding motifs or actinin type actin binding motifs. There are no N-myristoylation pattern or prenylation patterns.[25]
Kinase finder at Cuckoo determined kinase binding sites for SPMAP1. There are many Serine/Threonine, and Tyrosine kinase phosphorylation sites.[26] Serine and Threonine kinase binding sites are the most prevalent above the statistically significant threshold. There are no SUMOylation sites.[27] SPMAP1 gene has six sites on the sequence of possible O-GlcNAc sites.[28] Highly conserved O-GlcNAc amino acid sites are 24, 32, 117, and 142. O-GlcNAc post-translational modification occurs on Ser/Thr residues, specifically on oncogenes, tumor suppressors, and proteins involved in growth factor signaling.[29]
SPMAP1 has a Caspase3/7 motif, where either Caspase 3 or 7 would cleave.[30] This supports the idea that SPMAP1 is involved in proliferation, as a proapoptotic caspase would want to destroy any protein driving proliferation. The protein also has a motif where peptidyl-prolyl cis-trans isomerase NIMA interacting 1 (Pin1) binds.[30] Pin1 upregulation is involved in cancer and immune disorders.[31] This supports the claim that SPMAP1 is involved in cancer, immune cells, and perhaps cancers of the immune system. Additionally, SPMAP1 protein has an IBM site, where inhibitors of apoptosis (IAPs) bind.[30] This again supports the idea of SPMAP1 being involved in inhibiting apoptosis, and logically, driving cancer. Furthermore, SPMAP1 has motifs where GRB2's SH2 domain binds. GRB2 is an adapter protein involved in the RAS signaling pathway, a pathway that when deregulated drives uncontrolled proliferation.
Amino acid sequence
editA duplication may have occurred at positions 59–71.
Homo sapiens
MAYLSECRLRLEKGFILDGVAVSTAARAYGRSRPKLWSAIPPYNAQQDYHARSYFQ SHVVPPLLRVVPPLLRKTDQDHGGTGRDGWIVDYIHIFGQGQRYLNRRNWAGTGHS LQQVTGHDHYNADLKPIDGFNGRFGYRRNTPALRQSTSVFGEVTHFPLF
Associated proteins
editExpression
editProtein abundance in Homo sapiens whole organism is quite low. No data is available for other species.[36] Allen Brain Atlas yields no brain atlas for SPMAP1.[37]
Subcellular localization
editSPMAP1 protein has been found to be expressed in the intermediate filaments and the nucleoli.[38] A SPMAP1 antibody is available from Sigma-Aldrich.[39] Additionally, SPMAP1 localizes in the cytoplasm. Distantly related SPMAP1 orthologs in organisms such as Macrostomum lignano and Amphimedon queenslandica exhibit nuclear expression.[40] Nuclear localization signals are present in distantly related organisms in non-conserved sites. The results of the k-NN prediction is cytoplasmic localization.[41] SPMAP1 is not a signal peptide.[42] The protein is a soluble.[43]
Tissue
editLike most proteins, SPMAP1 protein is highly expressed in the testes.[44] The protein is expressed on adult tissues as well as fetal tissue. The protein has been found to be mildly expressed in connective tissue.[45] Additionally, expression has been seen in the sperm, breast epithelial cells, and various cells of the immune system.[46]
Clinical significance
editCancer
editProtein expression is elevated in many cancer patients. Specifically, protein expression has been shown to be high on colorectal, breast, prostate, and lung.[47] SPMAP1 is expressed in papillary thyroid cancer as well.[48] Additionally, mutations were found in SPMAP1 in endometrial, stomach, coloratura, and kidney cancer.[49] SPMAP1 expression is elevated in cancer patients with BRCA. In kidney renal clear cell carcinoma patients, SPMAP1 expression dramatically decreased compared to the non cancerous state.[13] In 80% of chromophobe renal cell carcinoma patients, at least one gene duplication SPMAP1 was present.[13]
Other conditions
editProtein expression is lower in males with teratozoospermia as compared to those without.[50] Many Geo Profile experiments have been conducted with SPMAP1, however, none yield data showing significant change in expression.[51]
Evolution
editSPMAP1 is a slow mutating protein. It resembles cytochrome c in its rate of divergence, as determined by the molecular clock equations.[52]
Paralogs
editThere are no known Homo sapiens paralogs for SPMAP1.[53]
Orthologs
editSPMAP1 protein has additional distantly related orthologs across the metazoan kingdom. Its most distant relative is in the sponge family. There is no known ortholog in ctenophores, nematodes, bacteria, fungus, plants, or zebrafish.[10] There are only two fish with the SPMAP1 gene. Model organisms such as Caenorhabditis elegans, and Drosophila melanogaster, do not have the gene.
SPMAP1 Orthologs[10]
| Sequence # | Genus and species | Common name | Accession # | Protein length | MYA Div | Seq Id | Confidence |
|---|---|---|---|---|---|---|---|
| 1 | Homo sapiens | Human | NP_001073934 | 154 | 0 | 100% | na |
| 2 | Camelus ferus | Wild Bactrian camel | XP_006176436 | 154 | 96 | 83% | 2.00E-94 |
| 3 | Pteropus alecto | Black flying fox | XP_006924784 | 154 | 96 | 81% | 1.00E-92 |
| 4 | Lipotes vexilifer | Yangtze river dolphin | XP_007465208 | 154 | 96 | 81% | 6.00E-89 |
| 5 | Condylura cristat | Star-nosed mole | XP_004684322 | 154 | 96 | 75% | 5.00E-78 |
| 6 | Myotis brandtii | Brandt's bat | EPQ05064 | 171 | 96 | 78% | 6.00E-78 |
| 7 | Marmata marmata marmata | Alpine marmot | XP_015362150.1 | 154 | 90 | 81% | 3.00E-94 |
| 8 | Octodon degus | Chilean rodent | XP_004633931 | 153 | 90 | 73% | 1.00E-76 |
| 9 | Alligator sinensis | Chinese alligator | XP_006022630 | 154 | 312 | 63% | 8.00E-68 |
| 10 | Anolis carolinensis | Lizard | XP_003222553 | 154 | 312 | 62% | 6.00E-67 |
| 11 | Xenopus laevis | African clawed frog | XP_018090228 | 244 | 352 | 51% | 4.00E-38 |
| 12 | Rhincodon typus | Whale shark | XP_020388051.1 | 164 | 476 | 53% | 5.00E-52 |
| 13 | Acanthaster planci | Starfish | XP_022086463 | 209 | 684 | 48% | 1.00E-37 |
| 14 | Mizuhopecten yessoensis | Scallop | XP_021340301 | 275 | 797 | 45% | 5.00E-06 |
| 15 | Lottia gigantea | Sea snail | XP_009063876 | 173 | 797 | 45% | 2.00E-37 |
| 16 | Lingula anatine | Lamp shell | XP_013388744.1 | 211 | 797 | 43% | 2.00E-35 |
| 17 | Biomphalaria glabrata | Freshwater snail | XP_013088317 | 198 | 797 | 41% | 6.00E-15 |
| 18 | Nematostella vectensis | Sea anemone | XP_001629616 | 173 | 824 | 48% | 2.00E-35 |
| 19 | Stylophora pistillata | Coral | XP_022795125 | 226 | 824 | 46% | 3.00E-38 |
| 20 | Macrostonum lignano | Flatworm | PAA73615 | 235 | 824 | 36% | 4.00E-25 |
| 21 | Amphimedon queenslandica | Sponge | XP_003389909 | 275 | 951.8 | 32% | 2.00E-12 |
References
edit- ^ a b c ENSG00000276913 GRCh38: Ensembl release 89: ENSG00000275489, ENSG00000276913 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000018543 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, et al. (April 2006). "DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage". Nature. 440 (7087): 1045–9. Bibcode:2006Natur.440.1045Z. doi:10.1038/nature04689. PMC 2610434. PMID 16625196.
- ^ PSORT II entry on c17orf98 https://psort.hgc.jp/form2.html
- ^ NCBI Conserved Domains entry C17orf98
- ^ a b ENMBL-EBI SAPS entry on c17orf98
- ^ "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2 May 2018.
- ^ a b c "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2 May 2018.
- ^ a b c Human protein atlas entry on c17orf98
- ^ Genomatix El Derado etnry on c17orf98
- ^ a b c TissGDB entry on c17orf98
- ^ Acieview entry on c17orf98
- ^ ClustalW entry on c17orf98 5' UTR
- ^ NCBI Blast entry on c17orf98 5' UTR https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blastho me
- ^ Genomatix El Derado etnry on c17orf98[permanent dead link]
- ^ Database, GeneCards Human Gene. "C17orf98 Gene - GeneCards - CQ098 Protein - CQ098 Antibody". www.genecards.org. Retrieved 2 May 2018.
- ^ "Genomatix El Derado etnry on c17orf98".[permanent dead link]
- ^ NCBI Genome Data Viewer
- ^ Target Scan entry on c17orf98 http://www.targetscan.org/cgibin/targetscan/vert_71/view_gene.cgi?rs=ENST00000398575.4&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1[permanent dead link]
- ^ Pax-db entry on c17orf98
- ^ "mFold entry on c17orf98 5' UTR".[permanent dead link]
- ^ ExPASy pI/mW entry on c17orf98 https://web.expasy.org/cgi-bin/compute_pi/pi_tool[permanent dead link]
- ^ PSort II entry on C17orf98[permanent dead link]
- ^ Bio Cockoo GPS entry on C17orf98 http://gps.biocu[permanent dead link]
- ^ GPS Sumo entry on c17orf98
- ^ YinOyang entry on c17orf98 http://www.cbs.dtu.dk/services/YinOYang/
- ^ Hanover, John A.; Krause, Michael W.; Love, Dona C. (2010). "The Hexosamine Signaling Pathway: O-GlcNAc cycling in feast or famine". Biochimica et Biophysica Acta (BBA) - General Subjects. 1800 (2): 80–95. doi:10.1016/j.bbagen.2009.07.017. PMC 2815088. PMID 19647043.
- ^ a b c Eukaryotic Linear Motif search on c17orf98 amino acid sequence
- ^ Esnault S, Braun RK, Shen ZJ, Xiang Z, Heninger E, Love RB, Sandor M, Malter JS (February 2007). "Pin1 modulates the type 1 immune response". PLOS ONE. 2 (2): e226. Bibcode:2007PLoSO...2..226E. doi:10.1371/journal.pone.0000226. PMC 1790862. PMID 17311089.
- ^ BioGrid entry on c17orf98
- ^ MINT entry on c17orf98
- ^ STRING entry on C17orf98
- ^ PSICQUIC View entry on c17orf98
- ^ pax-db entry on c17orf98 https://pax-db.org/protein/1858623#
- ^ "Microarray Data :: Allen Brain Atlas: Human Brain". human.brain-map.org. Retrieved 2018-05-06.
- ^ Human Protein Atlas (sigma) entry on c17orf98 https://www.proteinatlas.org/ENSG00000275489-C17orf98/cell[permanent dead link]
- ^ Sigma Aldrich entry on c17orf98 https://www.sigmaaldrich.com/catalog/product/sigma/hpa051696?lang=en®ion=US
- ^ PSORT II entry on c17orf98 amino acid sequence https://psort.hgc.jp/form2.html
- ^ PSort II entry on C17orf98 https://psort.hgc.jp/cgi-bin/runpsort.pl[permanent dead link]
- ^ DTU Bioinformatics entry on c17orf98
- ^ Expasy Sosui entry on C17orf98
- ^ Protein Atlas entry on c17orf98
- ^ NCBI Unigene entry on c17orf98 www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=169593&TAXID=9606&SEARCH=c17orf98
- ^ "Bio GPS entry on c17orf98".
- ^ Human Protein Atlas (sigma) entry on c17orf98 https://www.proteinatlas.org/ENSG00000275489-C17orf98/cell
- ^ NCBI GeoProfiles entry on c17orf98 https://www.ncbi.nlm.nih.gov/geoprofiles
- ^ Phosphosite entry on c17orf98 https://www.phosphosite.org/proteinAction.action?id=5156341&showAllSites=true
- ^ "C17orf98 - Teratozoospermia (HG-U133 2.0 )".
- ^ "NCBI GeoProfiles entry on c17orf98".
- ^ "The Molecular Clock and Estimating Species Divergence - Learn Science at Scitable". www.nature.com. Retrieved 2 May 2018.
- ^ Blast entry on c17orf98 https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins



