
Size of this PNG preview of this SVG file: 622 × 516 pixels. Other resolutions: 289 × 240 pixels | 579 × 480 pixels | 926 × 768 pixels | 1,234 × 1,024 pixels | 2,469 × 2,048 pixels.
Original file (SVG file, nominally 622 × 516 pixels, file size: 44 KB)
| This is a file from the Wikimedia Commons. Information from its description page there is shown below. Commons is a freely licensed media file repository. You can help. |
Summary
| DescriptionPathway Atrazine degradation.svg |
English: The s-triazine atrazine is a widely used herbicide that is persistent in soils. It is prone to ground and surface water contamination. A range of soil bacteria, including both Gram-negative and Gram-positive strains can degrade atrazine, utilizing it as a nitrogen and carbon source. In this aerobic pathway, which has been studied in most detail in the bacterium Pseudomonas sp. strain ADP, atrazine is catabolized in three enzymatic steps to cyanurate (a trimer), which can be further metabolized by ring cleavage to carbon dioxide and ammonia. The first hydrolytic enzyme, encoded by either atzA gene or trzN gene (EC 3.8.1.8), converts atrazine to hydroxy-atrazine. Two additional hydrolases, encoded by the atzB (EC 3.5.99.3) and atzC (EC 3.5.99.4) genes, continue the process by removing the ethylamine and isopropYl-amine groups. While some micro-organisms possess all of the required enzymes, others degrade atrazine by a community-approach, where different micro-organisms have some of the enzymes, and the intermediates in the metabolic pathway are passed between the soil bacteria.
References
|
||
| Date | |||
| Source | Own work | ||
| Author | J3D3 |
Licensing
I, the copyright holder of this work, hereby publish it under the following license:
This file is licensed under the Creative Commons Attribution-Share Alike 4.0 International license.
- You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
- Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- share alike – If you remix, transform, or build upon the material, you must distribute your contributions under the same or compatible license as the original.
Captions
Add a one-line explanation of what this file represents
Items portrayed in this file
depicts
7 September 2014
7 September 2014
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 04:24, 7 September 2014 | 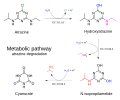 | 622 × 516 (44 KB) | J3D3 | User created page with UploadWizard |
File usage
The following page uses this file:
Global file usage
The following other wikis use this file:
Metadata
This file contains additional information, probably added from the digital camera or scanner used to create or digitize it.
If the file has been modified from its original state, some details may not fully reflect the modified file.
| Width | 621.51324 |
|---|---|
| Height | 516.13379 |
Retrieved from "https://en.wikipedia.org/wiki/File:Pathway_Atrazine_degradation.svg"