Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease.[1] Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.[2]
| Scytalidopepsin B | |||||||
|---|---|---|---|---|---|---|---|
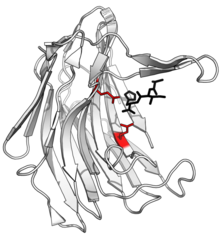 Structure of scytalidocarboxyl peptidase B, with cleaved peptide product in black and active site glutamate-glutamine dyad side chains in red. (PDB: 1S2K) | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | N/A | ||||||
| PDB | 1S2K | ||||||
| UniProt | P15369 | ||||||
| Other data | |||||||
| EC number | 3.4.23.32 | ||||||
| |||||||
Distribution and types
editThere are two independent families of glutamic proteases (G1 and G2), and have a limited distribution. They were originally thought to be limited to filamentous fungi mainly in the Ascomycota phylum.[3] Subsequently, however, glutamic proteases have been identified in bacteria and archaea.[4] A glutamic protease from a plant virus (strawberry mottle virus) has also been identified.[5]
The first superfamily of glutamic proteases was identified in the fungi Scytalidium lignicola and Aspergillus niger var. macrosporus, from which scytalidoglutamic peptidase (eqolisin) and aspergilloglutamic peptidase are derived respectively. These two proteases contain active site Glu and Gln residues and are grouped under MEROPS family G1.[6][7]
A convergently evolved glutamic peptidase, the pre-neck appendage protein (bacteriophage phi-29), uses a Glu and Asp dyad at the active site, and is classified as MEROPS family G2.[8]
Properties
editThese enzymes are acid proteases; eqolisin for example is most active at pH 2.0 when casein is used as substrate.[2] Eqolosins prefer bulky amino acid residues at the P1 site and small amino acid residues at the P1′ site. A characteristic of the protease is its insensitivity to pepstatin and S-PI (acetyl pepstatin) and it was previously classed as "pepstatin-insensitive carboxyl proteinases".[9] The other "pepstatin-insensitive carboxyl proteinases" belongs to subfamily of serine protease, serine-carboxyl protease (sedolisin) which was discovered in 2001.[2] These proteases are also not inhibited by DAN (diazoacetyl-DL-norleucine methylester) (7) but may be inhibited by EPNP (1,2-epoxy-3-(p-nitrophenoxy) propane).[10][11]
Active site and mechanism of catalysis
editThe active site of eqolosin contains a distinctive glutamic acid and glutamine catalytic dyad which are involved in substrate binding and catalysis. These residues act as a nucleophile, with the glutamic acid serving as a general acid in the first phase of the reaction, donating a proton to the carbonyl oxygen in the peptide bond of the substrate. One or two water molecules may be involved in the reaction supplying a hydroxyl group, and the glutamic acid further donates a proton to the amide nitrogen, resulting in breakage of the peptide bond. The glutamine then returns the glutamic acid to its initial state.[12]
See also
editReferences
edit- ^ Fujinaga M, Cherney MM, Oyama H, Oda K, James MN (Mar 2004). "The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum". Proceedings of the National Academy of Sciences of the United States of America. 101 (10): 3364–9. Bibcode:2004PNAS..101.3364F. doi:10.1073/pnas.0400246101. PMC 373467. PMID 14993599.
- ^ a b c Oda K (Jan 2012). "New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases". Journal of Biochemistry. 151 (1): 13–25. doi:10.1093/jb/mvr129. PMID 22016395.
- ^ Sims AH, Dunn-Coleman NS, Robson GD, Oliver SG (Oct 2004). "Glutamic protease distribution is limited to filamentous fungi". FEMS Microbiology Letters. 239 (1): 95–101. doi:10.1016/j.femsle.2004.08.023. PMID 15451106.
- ^ Jensen K, Østergaard PR, Wilting R, Lassen SF (2010). "Identification and characterization of a bacterial glutamic peptidase". BMC Biochemistry. 11 (47): 47. doi:10.1186/1471-2091-11-47. PMC 3009609. PMID 21122090.
- ^ Mann KS, Chisholm J, and Sanfaçon H (2019). "Strawberry Mottle Virus (Family Secoviridae, Order Picornavirales) Encodes a Novel Glutamic Protease To Process the RNA2 Polyprotein at Two Cleavage Sites". J Virol. 93 (5): e01679-18. doi:10.1128/JVI.01679-18. PMC 6384087. PMID 30541838.
- ^ Sasaki H, Kubota K, Lee WC, Ohtsuka J, Kojima M, Iwata S, Nakagawa A, Takahashi K, Tanokura M (Jul 2012). "The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis". Journal of Biochemistry. 152 (1): 45–52. doi:10.1093/jb/mvs050. PMID 22569035.
- ^ Takahashi K (2013). "Structure and function studies on enzymes with a catalytic carboxyl group(s): from ribonuclease T1 to carboxyl peptidases". Proceedings of the Japan Academy, Series B. 89 (6): 201–25. Bibcode:2013PJAB...89..201T. doi:10.2183/pjab.89.201. PMC 3749792. PMID 23759941.
- ^ "Family G2". MEROPS.[permanent dead link]
- ^ "Family G1". MEROPS.[permanent dead link]
- ^ Murao S, Oda K, Matsushita Y (1973). "Isolation and identification of a microorganism which produces non Streptomyces pepsin inhibitor and N-diazoacetyl-DL-norleucine methylester sensitive acid proteases". Agric. Biol. Chem. 37 (6): 1417–1421. doi:10.1271/bbb1961.37.1417.
- ^ Morihara K, Tsuzuki H, Murao S, Oda K (Mar 1979). "Pepstatin-insenstive acid proteases from Scytalidium lignicolum. Kinetic study with synthetic peptides". Journal of Biochemistry. 85 (3): 661–8. PMID 34596.
- ^ Moselio Schaechter, ed. (2009). Encyclopedia of Microbiology (3rd ed.). Academic Press. p. 499. ISBN 978-0123739391.