Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae of the family Straboviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains (or isolates), Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
| Escherichia virus T4 | |
|---|---|

| |
| Escherichia virus T4 (EM of virion) | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Uroviricota |
| Class: | Caudoviricetes |
| Family: | Straboviridae |
| Genus: | Tequatrovirus |
| Species: | Escherichia virus T4
|
| Strains[2] | |
| Synonyms[3] | |
|
Enterobacteria phage T4 | |
Use in research
editDating back to the 1940s and continuing today, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phage's genetic information is made up of around 300 genes. Coincident with their complexity, T-even viruses were found to have the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine.[4]
Genome and structure
editThe T4 virus's double-stranded DNA genome is about 169 kbp long[5] and encodes 289 proteins. The T4 genome is terminally redundant. Upon DNA replication, long multi-genome length concatemers are formed, perhaps by a rolling circle mechanism of replication.[6] When packaged, the concatemer is cut at unspecific positions of the same length, leading to several genomes that represent circular permutations of the original.[7] The T4 genome bears eukaryote-like intron sequences.
Translation
editThe Shine-Dalgarno sequence GAGG dominates in virus T4 early genes, whereas the sequence GGAG is a target for the T4 endonuclease RegB that initiates the early mRNA degradation.[8]
Virus particle structure
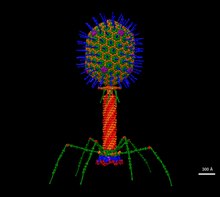
editT4 is a relatively large virus, at approximately 90 nm wide and 200 nm long (most viruses range from 25 to 200 nm in length). The DNA genome is held in an icosahedral head, also known as a capsid.[9] The T4's tail is hollow so that it can pass its nucleic acid into the cell it is infecting after attachment. Myoviridae phages like T4 have complex contractile tail structures with a large number of proteins involved in the tail assembly and function.[10] The tail fibres are also important in recognizing host cell surface receptors, so they determine if a bacterium is within the virus's host range.[11]
The structure of the 6 megadalton T4 baseplate that comprises 127 polypeptide chains of 13 different proteins (gene products 5, 5.4, 6, 7, 8, 9, 10, 11, 12, 25, 27, 48 and 53) has recently been described in atomic detail. An atomic model of the proximal region of the tail tube formed by gp54 and the main tube protein gp19 have also been created. The tape measure protein gp29 is present in the baseplate-tail tube complexes, but it could not be modeled.[12]
During assembly of the bacteriophage (phage) T4 virion, the morphogenetic proteins encoded by the phage genes interact with each other in a characteristic sequence. Maintaining an appropriate balance in the amounts of each of these proteins produced during viral infection appears to be critical for normal phage T4 morphogenesis.[13] Phage T4 encoded proteins that determine virion structure include major structural components, minor structural components and non-structural proteins that catalyze specific steps in the morphogenesis sequence.[14] Phage T4 morphogenesis is divided into three independent pathways: the head, the tail and the long tail fibres as detailed by Yap and Rossman.[15]
Infection process
editThe T4 virus initiates an Escherichia coli infection by binding OmpC porin proteins and lipopolysaccharide (LPS) on the surface of E. coli cells with its long tail fibers (LTF).[16][17] A recognition signal is sent through the LTFs to the baseplate. This unravels the short tail fibers (STF) that bind irreversibly to the E. coli cell surface. The baseplate changes conformation and the tail sheath contracts, causing GP5 at the end of the tail tube to puncture the outer membrane of the cell.[18] The lysozyme domain of GP5 is activated and degrades the periplasmic peptidoglycan layer. The remaining part of the membrane is degraded and then DNA from the head of the virus can travel through the tail tube and enter the E. coli cell.[citation needed]
In 1952, Hershey and Chase[19] provided key evidence that the phage DNA, as distinct from protein, enters the host bacterial cell upon infection and is thus the genetic material of the phage. This finding suggested that DNA is, in general, the genetic material of different organisms.[citation needed]
Reproduction
editThe lytic life cycle (from entering a bacterium to its destruction) takes approximately 30 minutes (at 37 °C). Virulent bacteriophages multiply in their bacterial host immediately after entry. After the number of progeny phages reach a certain amount, they cause the host to lyse or break down, therefore they would be released and infect new host cells.[20] The process of host lyses and release is called the lytic cycle. Lytic cycle is a cycle of viral reproduction that involves the destruction of the infected cell and its membrane. This cycle involves a virus that overtakes the host cell and its machinery to reproduce. Therefore, the virus must go through 5 stages in order to reproduce and infect the host cell:[citation needed]
- Adsorption and penetration (starting immediately)
- Arrest of host gene expression (starting immediately)
- Enzyme synthesis (starting after 5 minutes)
- DNA replication (starting after 10 minutes)
- Formation of new virus particles (starting after 12 minutes)
After the life cycle is complete, the host cell bursts open and ejects the newly built viruses into the environment, destroying the host cell. T4 has a burst size of approximately 100-150 viral particles per infected host.[citation needed]
Benzer (1955 – 1959) developed a system for studying the fine structure of the gene using bacteriophage T4 mutants defective in the rIIA and rIIB genes.[21][22][23] The techniques employed were complementation tests and crosses to detect recombination, particularly between deletion mutations. These genetic experiments led to the finding of a unique linear order of mutational sites within the genes. This result provided strong evidence for the key idea that the gene has a linear structure equivalent to a length of DNA with many sites that can independently mutate.[citation needed]
Adsorption and penetration
editJust like all other viruses, T-even phages do not randomly attach to the surface of their host; instead they "search" and bind to receptors, specific protein structures, found on the surface of the host. These receptors vary with the phage; teichoic acid, cell wall proteins and lipopolysaccharides, flagella, and pili all can serve as receptors for the phage to bind to. In order for the T-even phage to infect its host and begin its life cycle it must enter the first process of infection, adsorption of the phage to the bacterial cell. Adsorption is a value characteristic of phage-host pair and the adsorption of the phage on host cell surface is illustrated as a 2-stage process: reversible and irreversible. It involves the phages tail structure that begins when the phages tail fibers helps bind the phage to the appropriate receptor of its host. This process is reversible. One or more of the components of the base plate mediates irreversible process of binding of the phage to a bacterium.[citation needed]
Penetration is also a value characteristic of phage-host infection that involves the injection of the phages genetic material inside the bacterium. Penetration of nucleic acid takes place after the irreversible adsorption phase. Mechanisms involving penetration of the phages nucleic acid are specific for each phage. This penetration mechanism can involve electrochemical membrane potential, ATP molecules, enzymatic splitting of peptidoglycan layer, or all three of these factor can be vital for the penetration of the nucleic acid inside the bacterial cell. Studies have been done on the T2 bacteriophage (T4-like phage) mechanism of penetration and it has shown that the phage's tail does not penetrate inside the bacterial cell wall and penetration of this phage involves electrochemical membrane potential on the inner membrane.[24][25]
Replication and packaging
editVirus T4 genome is synthesized within the host cell using rolling circle replication.[6] The time it takes for DNA replication in a living cell was measured as the rate of virus T4 DNA elongation in virus-infected E. coli.[26] During the period of exponential DNA increase at 37 °C, the rate was 749 nucleotides per second. The mutation rate per base pair per replication during virus T4 DNA synthesis is 1.7 per 10−8,[27] a highly accurate DNA copying mechanism, with only 1 error in 300 copies. The virus also codes for unique DNA repair mechanisms.[28] The T4 phage head is assembled empty around a scaffolding protein, which is later degraded. Consequently, the DNA needs to enter the prohead through a tiny pore, which is achieved by a hexamer of gp17 interacting with DNA first, which also serves as a motor and nuclease. The T4 DNA packaging motor has been found to load DNA into virus capsids at a rate up to 2000 base pairs per second. The power involved, if scaled up in size, would be equivalent to that of an average automobile engine.[29]
Release
editThe final step in viral reproduction and multiplication is determined by the release of virions from the host cell. The release of the virions occurs after the breakage of the bacterial plasma membrane. Nonenveloped viruses lyse the host cell which is characterized by viral proteins attacking the peptidoglycan or membrane. The lysis of the bacteria occurs when the capsids inside the cell release the enzyme lysozyme which break down the cell wall. The released bacteriophages infect other cells, and the viral multiplication cycle is repeated within those cells.[citation needed]
Multiplicity reactivation
editMultiplicity reactivation (MR) is the process by which two or more virus genomes, each containing inactivating genome damage, can interact within an infected cell to form a viable virus genome. Salvador Luria, while studying UV irradiated virus T4 in 1946, discovered MR and proposed that the observed reactivation of damaged virus occurs by a recombination mechanism.(see refs.[30][31][32]) This preceded the confirmation of DNA as the genetic material in 1952 in related virus T2 by the Hershey–Chase experiment.[19]
As remembered by Luria (1984,[33] pg. 97) the discovery of reactivation of irradiated virus (referred to as "multiplicity reactivation") immediately started a flurry of activity in the study of repair of radiation damage within the early phage group (reviewed by Bernstein[28] in 1981). It turned out later that the repair of damaged virus by mutual help that Luria had discovered was only one special case of DNA repair. Cells of all types, not just, bacteria and their viruses, but all organisms studied, including humans, are now known to have complex biochemical processes for repairing DNA damages (see DNA repair). DNA repair processes are also now recognized as playing critical roles in protecting against aging, cancer, and infertility.[citation needed]
MR is usually represented by "survival curves" where survival of plaque forming ability of multiply infected cells (multicomplexes) is plotted against dose of genome damaging agent. For comparison, the survival of virus plaque forming ability of singly infected cells (monocomplexes) is also plotted against dose of genome damaging agent. The top figure shows the survival curves for virus T4 multicomplexes and monocomplexes with increasing dose of UV light. Since survival is plotted on a log scale it is clear that survival of multicomplexes exceeds that of monocomplexes by very large factors (depending on dose). The UV inactivation curve for multicomplexes has an initial shoulder. Other virus T4 DNA damaging agents with shoulders in their multicomplex survival curves are X-rays[34][35] and ethyl methane sulfonate (EMS).[28] The presence of a shoulder has been interpreted to mean that two recombinational processes are used.[36] The first one repairs DNA with high efficiency (in the "shoulder"), but is saturated in its ability as damage increases; the second pathway functions at all levels of damage. Surviving T4 virus released from multicomplexes show no increase in mutation, indicating that MR of UV irradiated virus is an accurate process.[36]
The bottom figure shows the survival curves for inactivation of virus T4 by the DNA damaging agent mitomycin C (MMC). In this case the survival curve for multicomplexes has no initial shoulder, suggesting that only the second recombinational repair process described above is active. The efficiency of repair by this process is indicated by the observation that a dose of MMC that allows survival of only 1 in 1,000 monocomplexes allows survival of about 70% of multicomplexes. Similar multicomplex survival curves (without shoulders) were also obtained for the DNA damaging agents P32 decay, psoralen plus near-UV irradiation (PUVA), N-methyl-N'-nitro-N-nitrosoguanidine (MNNG), methyl methane sulfonate (MMS) and nitrous acid.[28]
Several of the genes found to be necessary for MR in virus T4 proved to be orthologs for genes essential for recombination in prokaryotes, eukaryotes and archaea. This includes, for instance, T4 gene uvsX[37] which specifies a protein that has three-dimensional structural homology to RecA from Escherichia coli and the homologous protein RAD51 in eukaryotes and RadA in archaea. It has been suggested that the efficient and accurate recombinational repair of DNA damages during MR may be analogous to the recombinational repair process that occurs during meiosis in eukaryotes.[38]
History
editBacteriophages were first discovered by the English scientist Frederick Twort in 1915 and Félix d'Hérelle in 1917. In the late 1930s, T. L. Rakieten proposed either a mixture of raw sewerage or a lysate from E. coli infected with raw sewerage to the two researchers Milislav Demerec and Ugo Fano. These two researchers isolated T3, T4, T5, and T6 from E.coli. Also, in 1932, the researcher J. Bronfenbrenner had studied and worked on the T2 phage, at which the T2 phage was isolated from the virus.[39] This isolation was made from a fecal material rather than from sewerage. At any rate, Max Delbrück was involved in the discovery of the T even phages. His part was naming the bacteriophages into Type 1(T1), Type 2 (T2), Type 3 (T3), etc.[citation needed]
The specific time and place of T4 virus isolation remains unclear, though they were likely found in sewage or fecal material. T4 and similar viruses were described in a paper by Thomas F. Anderson, Max Delbrück, and Milislav Demerec in November 1944.[40] In 1943, Salvador Luria and Delbrück showed that bacterial mutations for phage resistance arise in the absence of selection, rather than being a response to selection.[33] The traditional wisdom among bacteriologists prior to 1943 was that bacteria had no chromosomes and no genes. The Luria–Delbrück experiment showed that bacteria, like other established model genetic organisms, have genes, and that these can spontaneously mutate to generate mutants that may then reproduce to form clonal lineages. That year, they also began working with Alfred Hershey, another phage experimenter.[41] (The three would share the 1969 Nobel Prize in Physiology or Medicine, "for work on the replication mechanism and genetics of viruses".)
The phage group was an informal network of biologists centered on Max Delbrück that carried out basic research mainly on bacteriophage T4 and made numerous seminal contributions to microbial genetics and the origins of molecular biology in the mid-20th century. In 1961, Sydney Brenner, an early member of the phage group, collaborated with Francis Crick, Leslie Barnett and Richard Watts-Tobin at the Cavendish Laboratory in Cambridge to perform genetic experiments that demonstrated the basic nature of the genetic code for proteins.[42] These experiments, carried out with mutants of the rIIB gene of phage T4, showed, that for a gene that encodes a protein, three sequential bases of the gene's DNA specify each successive amino acid of the protein. Thus the genetic code is a triplet code, where each triplet (called a codon) specifies a particular amino acid. They also obtained evidence that the codons do not overlap with each other in the DNA sequence encoding a protein, and that such a sequence is read from a fixed starting point.[citation needed]
During 1962-1964 phage T4 researchers provided an opportunity to study the function of virtually all of the genes that are essential for growth of the phage under laboratory conditions.[43][44] These studies were facilitated by the discovery of two classes of conditional lethal mutants. One class of such mutants is known as amber mutants.[45] Another class of conditional lethal mutants is referred to as temperature-sensitive mutants[46] Studies of these two classes of mutants led to considerable insight into numerous fundamental biologic problems. Thus understanding was gained on the functions and interactions of the proteins employed in the machinery of DNA replication, repair and recombination, and on how viruses are assembled from protein and nucleic acid components (molecular morphogenesis). Furthermore, the role of chain terminating codons was elucidated. One noteworthy study used amber mutants defective in the gene encoding the major head protein of phage T4.[47] This experiment provided strong evidence for the widely held, but prior to 1964 still unproven, "sequence hypothesis" that the amino acid sequence of a protein is specified by the nucleotide sequence of the gene determining the protein. Thus, this study demonstrated the co-linearity of the gene with its encoded protein.
A number of Nobel Prize winners worked with virus T4 or T4-like viruses including Max Delbrück, Salvador Luria, Alfred Hershey, James D. Watson, and Francis Crick. Other important scientists who worked with virus T4 include Michael Rossmann, Seymour Benzer, Bruce Alberts, Gisela Mosig,[48] Richard Lenski, and James Bull.
See also
editReferences
edit- ^ Padilla-Sanchez V (2021). "Structural Model of Bacteriophage T4". WikiJournal of Science. 4 (1): 5. doi:10.15347/WJS/2021.005.
- ^ "ICTV 9th Report (2011) Myoviridae". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 26 December 2018. Retrieved 26 December 2018.
- ^ "ICTV Taxonomy history: Escherichia virus T4". International Committee on Taxonomy of Viruses (ICTV). Retrieved 26 December 2018.
Caudovirales > Myoviridae > Tevenvirinae > T4virus > Escherichia virus T4
- ^ Wyatt GR, Cohen SS (December 1952). "A New Pyrimidine Base from Bacteriophage Nucleic Acids". Nature. 170 (4338): 1072–1073. Bibcode:1952Natur.170.1072W. doi:10.1038/1701072a0. ISSN 1476-4687. PMID 13013321. S2CID 4277592.
- ^ Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W (March 2003). "Bacteriophage T4 genome". Microbiology and Molecular Biology Reviews. 67 (1): 86–156, table of contents. doi:10.1128/mmbr.67.1.86-156.2003. PMC 150520. PMID 12626685.
- ^ a b Bernstein H, Bernstein C (July 1973). "Circular and branched circular concatenates as possible intermediates in bacteriophage T4 DNA replication". Journal of Molecular Biology. 77 (3): 355–61. doi:10.1016/0022-2836(73)90443-9. PMID 4580243.
- ^ Madigan M, Martinko J, eds. (2006). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 978-0-13-144329-7.
- ^ Malys N (January 2012). "Shine-Dalgarno sequence of bacteriophage T4: GAGG prevails in early genes". Molecular Biology Reports. 39 (1): 33–9. doi:10.1007/s11033-011-0707-4. PMID 21533668. S2CID 17854788.
- ^ Prescott LM, Harley JP, Klein DA (2008). Microbiology (seventh ed.). McGraw-Hill. ISBN 978-0-07-126727-4.
- ^ Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG (December 2010). "Morphogenesis of the T4 tail and tail fibers". Virology Journal. 7: 355. doi:10.1186/1743-422X-7-355. PMC 3004832. PMID 21129200.
- ^ Ackermann HW, Krisch HM (1997). "A catalogue of T4-type bacteriophages". Archives of Virology. 142 (12): 2329–45. doi:10.1007/s007050050246. PMID 9672598. S2CID 39369249.
- ^ Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG (May 2016). "Structure of the T4 baseplate and its function in triggering sheath contraction". Nature. 533 (7603): 346–52. Bibcode:2016Natur.533..346T. doi:10.1038/nature17971. PMID 27193680. S2CID 4399265.
- ^ Floor E (February 1970). "Interaction of morphogenetic genes of bacteriophage T4". Journal of Molecular Biology. 47 (3): 293–306. doi:10.1016/0022-2836(70)90303-7. PMID 4907266.
- ^ Snustad DP (August 1968). "Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric". Virology. 35 (4): 550–63. doi:10.1016/0042-6822(68)90285-7. PMID 4878023.
- ^ Yap ML, Rossmann MG (2014). "Structure and function of bacteriophage T4". Future Microbiology. 9 (12): 1319–27. doi:10.2217/fmb.14.91. PMC 4275845. PMID 25517898.
- ^ Yu F, Mizushima S (August 1982). "Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4". Journal of Bacteriology. 151 (2): 718–22. doi:10.1128/JB.151.2.718-722.1982. PMC 220313. PMID 7047495.
- ^ Furukawa H, Mizushima S (May 1982). "Roles of cell surface components of Escherichia coli K-12 in bacteriophage T4 infection: interaction of tail core with phospholipids". Journal of Bacteriology. 150 (2): 916–24. doi:10.1128/JB.150.2.916-924.1982. PMC 216445. PMID 7040345.
- ^ Maghsoodi A, Chatterjee A, Andricioaei I, Perkins NC (December 2019). "How the phage T4 injection machinery works including energetics, forces, and dynamic pathway". Proceedings of the National Academy of Sciences of the United States of America. 116 (50): 25097–25105. Bibcode:2019PNAS..11625097M. doi:10.1073/pnas.1909298116. PMC 6911207. PMID 31767752.
- ^ a b HERSHEY AD, CHASE M (May 1952). "Independent functions of viral protein and nucleic acid in growth of bacteriophage". The Journal of General Physiology. 36 (1): 39–56. doi:10.1085/jgp.36.1.39. PMC 2147348. PMID 12981234.
- ^ Sherwood L (2011). Prescott's Microbiology (eighth ed.). McGraw-Hill.
- ^ Benzer S. "Adventures in the rII region" in Phage and the Origins of Molecular Biology (2007) Edited by John Cairns, Gunther S. Stent, and James D. Watson, Cold Spring Harbor Laboratory of Quantitative Biology, Cold Spring Harbor, Long Island, New York ISBN 978-0879698003
- ^ Benzer S (June 1955). "Fine structure of a genetic region in bacteriophage". Proceedings of the National Academy of Sciences of the United States of America. 41 (6): 344–54. Bibcode:1955PNAS...41..344B. doi:10.1073/pnas.41.6.344. PMC 528093. PMID 16589677.
- ^ Benzer S (November 1959). "On the topology of genetic fine structure". Proceedings of the National Academy of Sciences of the United States of America. 45 (11): 1607–20. Bibcode:1959PNAS...45.1607B. doi:10.1073/pnas.45.11.1607. PMC 222769. PMID 16590553.
- ^ Norkin LC (2010). Virology, Molecular Biology and Pathogenesis. Washington: American Society for Microbiology. p. 31. ISBN 978-1-55581-453-3.
- ^ Prescott LM, Harley JP, Klein DA (2008). Microbiology (seventh ed.). McGraw Hill. p. 427. ISBN 978-0-07-126727-4.
- ^ McCarthy D, Minner C, Bernstein H, Bernstein C (1976). "DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant". J Mol Biol. 106 (4): 963–81. doi:10.1016/0022-2836(76)90346-6. PMID 789903.
- ^ Drake JW (1970) The Molecular Basis of Mutation. Holden-Day, San Francisco ISBN 0816224501 ISBN 978-0816224500
- ^ a b c d Bernstein C. "Deoxyribonucleic acid repair in bacteriophage". Microbiol Rev. 1981 Mar;45(1):72-98. Review. PMID 6261109
- ^ Rao VB, Black LW (December 2010). "Structure and assembly of bacteriophage T4 head". Virology Journal. 7: 356. doi:10.1186/1743-422X-7-356. PMC 3012670. PMID 21129201.
- ^ Luria SE (1947). "Reactivation of Irradiated Bacteriophage by Transfer of Self-Reproducing Units". Proc. Natl. Acad. Sci. U.S.A. 33 (9): 253–64. Bibcode:1947PNAS...33..253L. doi:10.1073/pnas.33.9.253. PMC 1079044. PMID 16588748.
- ^ LURIA SE, DULBECCO R (1948). "Lethal mutations, and inactivation of individual genetic determinants in bacteriophage". Genetics. 33 (6): 618. PMID 18100306.
- ^ Luria SE, Dulbecco R (1949). "Genetic Recombinations Leading to Production of Active Bacteriophage from Ultraviolet Inactivated Bacteriophage Particles". Genetics. 34 (2): 93–125. doi:10.1093/genetics/34.2.93. PMC 1209443. PMID 17247312.
- ^ a b Salvador E. Luria. A Slot Machine, A Broken Test Tube: An Autobiography. Harper & Row, New York: 1984. Pp. 228. ISBN 0-06-015260-5 (USA and Canada)
- ^ WATSON JD (1952). "The properties of x-ray inactivated bacteriophage". J. Bacteriol. 63 (4): 473–85. doi:10.1128/JB.63.4.473-485.1952. PMC 169298. PMID 14938320.
- ^ HARM W (1958). "Multiplicity reactivation, marker rescue, and genetic recombination in phage T4 following x-ray inactivation". Virology. 5 (2): 337–61. doi:10.1016/0042-6822(58)90027-8. PMID 13544109.
- ^ a b Yarosh DB (1978). "UV-induced mutation in bacteriophage T4". J. Virol. 26 (2): 265–71. doi:10.1128/JVI.26.2.265-271.1978. PMC 354064. PMID 660716.
- ^ Story RM, Bishop DK, Kleckner N, Steitz TA (1993). "Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast". Science. 259 (5103): 1892–6. Bibcode:1993Sci...259.1892S. doi:10.1126/science.8456313. PMID 8456313.
- ^ Bernstein C (1979). "Why are babies young? Meiosis may prevent aging of the germ line". Perspect. Biol. Med. 22 (4): 539–44. doi:10.1353/pbm.1979.0041. PMID 573881. S2CID 38550472.
- ^ Willey J. Prescott's Microbiology (seventh ed.). McGraw-Hill.
- ^ Abedon ST (June 2000). "The murky origin of Snow White and her T-even dwarfs". Genetics. 155 (2): 481–6. doi:10.1093/genetics/155.2.481. PMC 1461100. PMID 10835374.
- ^ Morange, A History of Molecular Biology, pp 43-44
- ^ CRICK FH, BARNETT L, BRENNER S, WATTS-TOBIN RJ (December 1961). "General nature of the genetic code for proteins". Nature. 192 (4809): 1227–32. Bibcode:1961Natur.192.1227C. doi:10.1038/1921227a0. PMID 13882203. S2CID 4276146.
- ^ Edgar RS Conditional lethals: in Phage and the Origins of Molecular Biology (2007) Edited by John Cairns, Gunther S. Stent, and James D. Watson, Cold Spring Harbor Laboratory of Quantitative Biology, Cold Spring Harbor, Long Island, New York ISBN 978-0879698003
- ^ Edgar B (October 2004). "The genome of bacteriophage T4: an archeological dig". Genetics. 168 (2): 575–82. doi:10.1093/genetics/168.2.575. PMC 1448817. PMID 15514035.
- ^ Epstein RH, Bolle A, Steinberg CM, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar RS, Susman M, Denhardt GH, Lielausis A (1963). "Physiological Studies of Conditional Lethal Mutants of Bacteriophage T4D". Cold Spring Harbor Symposia on Quantitative Biology. 28: 375–394. doi:10.1101/SQB.1963.028.01.053. ISSN 0091-7451.
- ^ Edgar RS, Lielausis I (April 1964). "Temperature-sensitive mutants of bacteriophage T4D: Their isolation and Characterization". Genetics. 49 (4): 649–62. doi:10.1093/genetics/49.4.649. PMC 1210603. PMID 14156925.
- ^ Sarabhai AS, Stretton AO, Brenner S, Bolle A (January 1964). "Co-linearity of the gene with the polypeptide chain". Nature. 201 (4914): 13–7. Bibcode:1964Natur.201...13S. doi:10.1038/201013a0. PMID 14085558. S2CID 10179456.
- ^ Nossal NG, Franklin JL, Kutter E, Drake JW (November 2004). "Anecdotal, historical and critical commentaries on genetics. Gisela Mosig". Genetics. 168 (3): 1097–104. doi:10.1093/genetics/168.3.1097. PMC 1448779. PMID 15579671.
Further reading
edit- Leiman P.G., Kanamaru S, Mesyanzhinov V.V., Arisaka F., Rossmann M.G. (2003). "Structure and morphogenesis of bacteriophage T4". Cellular and Molecular Life Sciences. 60 (11): 2356–2370. doi:10.1007/s00018-003-3072-1. PMC 11138918. PMID 14625682. S2CID 2228357.
- Karam, J., Petrov, V., Nolan, J., Chin, D., Shatley, C., Krisch, H., and Letarov, A. The T4-like phages genome project. https://web.archive.org/web/20070523215704/http://phage.bioc.tulane.edu/. (The T4-like phage full genomic sequence depository)
- Mosig, G., and F. Eiserling. 2006. T4 and related phages: structure and development, R. Calendar and S. T. Abedon (eds.), The Bacteriophages. Oxford University Press, Oxford. (Review of phage T4 biology) ISBN 0-19-514850-9
- Filee J. Tetart F., Suttle C.A., Krisch H.M. (2005). "Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere". Proc. Natl. Acad. Sci. USA. 102 (35): 12471–6. Bibcode:2005PNAS..10212471F. doi:10.1073/pnas.0503404102. PMC 1194919. PMID 16116082. (Indication of prevalence and T4-like phages in the wild)
- Chibani-Chennoufi S., Canchaya C., Bruttin A., Brussow H. (2004). "Comparative genomics of the T4-Like Escherichia coli phage JS98: implications for the evolution of T4 phages". J. Bacteriol. 186 (24): 8276–86. doi:10.1128/JB.186.24.8276-8286.2004. PMC 532421. PMID 15576776. (Characterization of a T4-like phage)
- Desplats C, Krisch HM (May 2003). "The diversity and evolution of the T4-type bacteriophages". Res. Microbiol. 154 (4): 259–67. doi:10.1016/S0923-2508(03)00069-X. PMID 12798230.
- Miller, E.S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W. (2003). "Bacteriophage T4 genome". Microbiol. Mol. Biol. Rev. 67 (1): 86–156. doi:10.1128/MMBR.67.1.86-156.2003. PMC 150520. PMID 12626685. (Review of phage T4, from the perspective of its genome)
- Desplats C., Dez C., Tetart F., Eleaume H., Krisch H.M. (2002). "Snapshot of the genome of the pseudo-T-even bacteriophage RB49". J. Bacteriol. 184 (10): 2789–2804. doi:10.1128/JB.184.10.2789-2804.2002. PMC 135041. PMID 11976309. (Overview of the RB49 genome, a T4-like phage)
- Malys N, Chang DY, Baumann RG, Xie D, Black LW (2002). "A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction". J Mol Biol. 319 (2): 289–304. doi:10.1016/S0022-2836(02)00298-X. PMID 12051907. (T4 phage application in biotechnology for studying protein interaction)
- Tétart F., Desplats C., Kutateladze M., Monod C., Ackermann H.-W., Krisch H.M. (2001). "Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages". J. Bacteriol. 183 (1): 358–366. doi:10.1128/JB.183.1.358-366.2001. PMC 94885. PMID 11114936. (Indication of the prevalence of T4-type sequences in the wild)
- Abedon S.T. (2000). "The murky origin of Snow White and her T-even dwarfs". Genetics. 155 (2): 481–6. doi:10.1093/genetics/155.2.481. PMC 1461100. PMID 10835374. (Historical description of the isolation of the T4-like phages T2, T4, and T6)
- Ackermann HW, Krisch HM (1997). "A catalogue of T4-type bacteriophages". Arch. Virol. 142 (12): 2329–45. doi:10.1007/s007050050246. PMID 9672598. S2CID 39369249. Archived from the original on 1 November 2001. (Nearly complete list of then-known T4-like phages)
- Monod C, Repoila F, Kutateladze M, Tétart F, Krisch HM (March 1997). "The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4". J. Mol. Biol. 267 (2): 237–49. doi:10.1006/jmbi.1996.0867. PMID 9096222. (Overview of various T4-like phages from the perspective of their genomes)
- Kutter E., Gachechiladze K., Poglazov A., Marusich E., Shneider M., Aronsson P., Napuli A., Porter D., Mesyanzhinov V. (1995). "Evolution of T4-related phages". Virus Genes. 11 (2–3): 285–297. doi:10.1007/BF01728666. PMID 8828153. S2CID 20529415. (Comparison of the genomes of various T4-like phages)
- Karam, J. D. et al. 1994. Molecular Biology of Bacteriophage T4. ASM Press, Washington, DC. (The second T4 bible, go here, as well as Mosig and Eiserling, 2006, to begin to learn about the biology T4 phage) ISBN 1-55581-064-0
- Eddy, S. R. 1992. Introns in the T-Even Bacteriophages. PhD thesis. University of Colorado at Boulder. (Chapter 3 provides overview of various T4-like phages as well as the isolation of then-new T4-like phages)
- Surdis, T.J "et al" UC Santa Cruz, Nov 1978, "Bacteriophage attachment methods specific to T4", analysis, Overview.
- Mathews, C. K., E. M. Kutter, G. Mosig, and P. B. Berget. 1983. Bacteriophage T4. American Society for Microbiology, Washington, DC. (The first T4 bible; not all information here is duplicated in Karam et al., 1994; see especially the introductory chapter by Doermann for a historical overview of the T4-like phages) ISBN 0-914826-56-5
- Russell, R. L. 1967. Speciation Among the T-Even Bacteriophages. PhD thesis. California Institute of Technology. (Isolation of the RB series of T4-like phages)
- Malys N, Nivinskas R (2009). "Non-canonical RNA arrangement in T4-even phages: accommodated ribosome binding site at the gene 26-25 intercistronic junction". Mol Microbiol. 73 (6): 1115–1127. doi:10.1111/j.1365-2958.2009.06840.x. PMID 19708923. S2CID 8187771. (rare type of translational regulation characterized in T4)
- Kay D., Fildes P. (1962). "Hydroxymethylcytosine-containing and tryptophan-dependent bacteriophages isolated from city effluents". J. Gen. Microbiol. 27: 143–6. doi:10.1099/00221287-27-1-143. PMID 14454648. (T4-like phage isolation, including that of phage Ox2)