The autoimmune regulator (AIRE) is a protein that in humans is encoded by the AIRE gene.[5] It is a 13kbp gene on chromosome 21q22.3 that encodes 545 amino acids.[6] AIRE is a transcription factor expressed in the medulla (inner part) of the thymus. It is part of the mechanism which eliminates self-reactive T cells that would cause autoimmune disease. It exposes T cells to normal, healthy proteins from all parts of the body, and T cells that react to those proteins are destroyed.
Each T cell recognizes a specific antigen when it is presented in complex with a major histocompatibility complex (MHC) molecule by an antigen presenting cell. This recognition is accomplished by the T cell receptors expressed on the cell surface. T cells receptors are generated by randomly shuffled gene segments which results in a highly diverse population of T cells—each with a unique antigen specificity. Subsequently, T cells with receptors that recognize the body's own proteins need to be eliminated while still in the thymus. Through the action of AIRE, medullary thymic epithelial cells (mTEC) express major proteins from elsewhere in the body (so called "tissue-restricted antigens" - TRA) and T cells that respond to those proteins are eliminated through cell death (apoptosis). Thus AIRE drives negative selection of self-recognizing T cells.[7] When AIRE is defective, T cells that recognize antigens normally produced by the body can exit the thymus and enter circulation. This can result in a variety of autoimmune diseases.
The gene was first reported by two independent research groups Aaltonen et al. and Nagamine et al. in 1997 who were able to isolate and clone the gene from human chromosome 21q22.3. Their work was able to show that mutations in the AIRE gene are responsible for the pathogenesis of Autoimmune polyglandular syndrome type I.[5][8] More insight into the AIRE protein was later provided by Heino et al. in 2000. They showed that AIRE protein is mainly expressed in the thymic medullary epithelial cells using immunohistochemistry.[9]
Function
editIn the thymus, the AIRE causes transcription of a wide selection of organ-specific genes that create proteins that are usually only expressed in peripheral tissues, creating an "immunological self-shadow" in the thymus.[10][11] It is important that self-reactive T cells that bind strongly to self-antigen are eliminated in the thymus (via the process of negative selection), otherwise they may later encounter and bind to their corresponding self-antigens and initiate an autoimmune reaction. So the expression of non-local proteins by AIRE in the thymus reduces the threat of autoimmunity by promoting the elimination of auto-reactive T cells that bind antigens not normally found in the thymus. Furthermore, it has been found that AIRE is expressed in a population of stromal cells located in secondary lymphoid tissues, however these cells appear to express a distinct set of TRAs compared to mTECs.[12]
Research in knockout mice has demonstrated that AIRE functions through initiating the transcription of a diverse set of self-antigens, such as insulin, in the thymus.[10] This expression then allows maturing thymocytes to become tolerant towards peripheral organs, thereby suppressing autoimmune disease.[11]
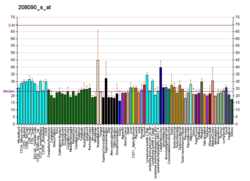
The AIRE gene is expressed in many other tissues as well.[13] The AIRE gene is also expressed in the 33D1+ subset of dendritic cells in mouse and in human dendritic cells.[14]
Structure
editAIRE is composed of a multidomain structure that is able to bind to chromatin and act as a regulator of gene transcription. The specific makeup of AIRE includes a caspase activation and recruitment domain (CARD), nuclear localization signal (NLS), SAND domain, and two plant-homeodomain (PHD) fingers.[15] The SAND domain is located in the middle of the amino-acid chain (aa 180-280) and mediates the binding of AIRE to phosphate groups of DNA.[16] Another potential role for this domain is to anchor AIRE to heterologous proteins.[17] The two cysteine-rich PHD finger domains at the C-terminus of AIRE are PHD1 (aa 299-340) and PHD2 (aa 434-475) which are separated by a proline-rich region of amino acids.[18] These finger domains serve to read chromatin marks through the degree of methylation at the tail of histone H3. More specifically, PHD1 is able to recognize unmethylation at the H3 tail as an epigenetic mark.[19]
An integral characteristic of AIRE is its ability to homomerize into dimers and trimers which allows it to bind to specific oligonucleotide motifs.[20] This property comes from the homogeneously staining region (HSR) located at the N-terminus. Because of the α-helical four-helix bundle structure, HSR’s are sensitive to conformational changes of the gene.[21] Variants and deletions involving this domain cause an inability to activate gene transcription by preventing oligomer formation and can result in APS-1.
Mechanism
editInstead of binding to consensus sequences of target gene promoters, like conventional transcription factors, AIRE engages in coordinated sequences that are performed by its multimolecular complexes. The first AIRE partner that was identified is the CREB-binding protein (CBP) that is localized in nuclear bodies and is a co-activator of many transcription factors.[21] Other AIRE partners include positive transcription elongation factor b (P-TEFb) and DNA activated protein kinase (DNA-PK).[22][23] DNA-PK phosphorylates AIRE in vitro at Thr68 and Ser156.[23] Another partner is DNA-topoisomerase (DNA-TOP) IIα. This isomerase enzyme works on DNA topology and removes positive and negative DNA supercoils by causing transient DNA breaks. In turn, this causes relaxation of local chromatin and helps the initiation and post-initiation events of gene transcription.[24] By performing double-stranded DNA breaks, DNA-TOPIIα recruits DNA-PK and poly-(ADP-ribose) polymerase (PARP1) which are involved in DNA break and repair through non-homologous end joining.[25]
Pathology
editThe AIRE gene is mutated in the rare autoimmune syndrome autoimmune polyendocrinopathy syndrome type 1 (APS-1), also known as autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). Different mutations are more common among certain populations in the world.[26] The most common exonic mutations of AIRE occur on exons 1, 2, 6, 8, and 10. Exons 1 and 2 encode the HSR, exon 6 encodes the SAND domain, exon 8 is in the PHD-1 domain, and exon 10 is located in the proline-rich region between the two PHD finger domains.[27] Known mutations in AIRE include Arg139X, Arg257X, and Leu323SerfsX51.[28]
Disruption of AIRE results in the development of a range of autoimmune diseases, the most common clinical conditions in the syndrome are hypoparathyroidism, primary adrenocortical failure and chronic mucocutaneous candidiasis.[29]
A gene knockout of the murine homolog of Aire has created a transgenic mouse model that is used to study the mechanism of disease in human patients.[30]
Interactions
editAutoimmune regulator has been shown to interact with CREB binding protein.[21][31]
See also
edit- List of human clusters of differentiation for a list of CD molecules
- Immune system
- Immune tolerance
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000160224 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000000731 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Aaltonen J, Björses P, Perheentupa J, Horelli-Kuitunen N, Palotie A, Peltonen L, et al. (Finnish-German APECED Consortium) (December 1997). "An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains". Nature Genetics. 17 (4): 399–403. doi:10.1038/ng1297-399. PMID 9398840. S2CID 29785642.
- ^ Blechschmidt K, Schweiger M, Wertz K, Poulson R, Christensen HM, Rosenthal A, et al. (February 1999). "The mouse Aire gene: comparative genomic sequencing, gene organization, and expression". Genome Research. 9 (2). Cold Spring Harbor Laboratory Press: 158–66. doi:10.1101/gr.9.2.158. OCLC 678392077. PMC 310712. PMID 10022980.
- ^ Anderson MS, Su MA (April 2011). "Aire and T cell development". Current Opinion in Immunology. 23 (2): 198–206. doi:10.1016/j.coi.2010.11.007. PMC 3073725. PMID 21163636.
- ^ Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. (December 1997). "Positional cloning of the APECED gene". Nature Genetics. 17 (4): 393–8. doi:10.1038/ng1297-393. PMID 9398839. S2CID 1583134.
- ^ Heino M, Peterson P, Sillanpää N, Guérin S, Wu L, Anderson G, et al. (July 2000). "RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse". European Journal of Immunology. 30 (7): 1884–93. doi:10.1002/1521-4141(200007)30:7<1884::aid-immu1884>3.0.co;2-p. PMID 10940877.
- ^ a b Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. (November 2002). "Projection of an immunological self shadow within the thymus by the aire protein". Science. 298 (5597): 1395–401. Bibcode:2002Sci...298.1395A. doi:10.1126/science.1075958. PMID 12376594. S2CID 13989491.
- ^ a b Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC (April 2003). "Aire regulates negative selection of organ-specific T cells". Nature Immunology. 4 (4): 350–4. doi:10.1038/ni906. PMID 12612579. S2CID 4561402.
- ^ Gardner JM, Devoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, et al. (August 2008). "Deletional tolerance mediated by extrathymic Aire-expressing cells". Science. 321 (5890): 843–7. Bibcode:2008Sci...321..843G. doi:10.1126/science.1159407. PMC 2532844. PMID 18687966.
- ^ "AIRE Gene expression/activity chart". BioGPS - your Gene Portal System. Archived from the original on 2009-12-30. Retrieved 2009-12-19.
- ^ Lindmark E, Chen Y, Georgoudaki AM, Dudziak D, Lindh E, Adams WC, et al. (May 2013). "AIRE expressing marginal zone dendritic cells balances adaptive immunity and T-follicular helper cell recruitment". Journal of Autoimmunity. 42: 62–70. doi:10.1016/j.jaut.2012.11.004. hdl:10616/41469. PMID 23265639.
- ^ Perniola R, Musco G (February 2014). "The biophysical and biochemical properties of the autoimmune regulator (AIRE) protein". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1842 (2): 326–37. doi:10.1016/j.bbadis.2013.11.020. PMID 24275490.
- ^ Gibson TJ, Ramu C, Gemünd C, Aasland R (July 1998). "The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor". Trends in Biochemical Sciences. 23 (7): 242–4. doi:10.1016/s0968-0004(98)01231-6. PMID 9697411.
- ^ Carles CC, Fletcher JC (July 2010). "Missing links between histones and RNA Pol II arising from SAND?". Epigenetics. 5 (5): 381–5. doi:10.4161/epi.5.5.11956. PMID 20458168. S2CID 42505863.
- ^ Aasland R, Gibson TJ, Stewart AF (February 1995). "The PHD finger: implications for chromatin-mediated transcriptional regulation". Trends in Biochemical Sciences. 20 (2): 56–9. doi:10.1016/s0968-0004(00)88957-4. PMID 7701562.
- ^ Org T, Chignola F, Hetényi C, Gaetani M, Rebane A, Liiv I, et al. (April 2008). "The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression". EMBO Reports. 9 (4): 370–6. doi:10.1038/embor.2008.11. PMC 2261226. PMID 18292755. S2CID 84265877.
- ^ Kumar PG, Laloraya M, Wang CY, Ruan QG, Davoodi-Semiromi A, Kao KJ, She JX (November 2001). "The autoimmune regulator (AIRE) is a DNA-binding protein". The Journal of Biological Chemistry. 276 (44): 41357–64. doi:10.1074/jbc.M104898200. PMID 11533054. S2CID 27962035.
- ^ a b c Pitkänen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, et al. (June 2000). "The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein". The Journal of Biological Chemistry. 275 (22): 16802–9. doi:10.1074/jbc.m908944199. PMID 10748110. S2CID 2518676.
- ^ Oven I, Brdicková N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM (December 2007). "AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells". Molecular and Cellular Biology. 27 (24): 8815–23. doi:10.1128/MCB.01085-07. OCLC 456127729. PMC 2169392. PMID 17938200.
- ^ a b Liiv I, Rebane A, Org T, Saare M, Maslovskaja J, Kisand K, et al. (January 2008). "DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1783 (1): 74–83. doi:10.1016/j.bbamcr.2007.09.003. PMC 2225445. PMID 17997173.
- ^ Pommier Y, Sun Y, Huang SN, Nitiss JL (November 2016). "Roles of eukaryotic topoisomerases in transcription, replication and genomic stability". Nature Reviews. Molecular Cell Biology. 17 (11): 703–721. doi:10.1038/nrm.2016.111. PMC 9248348. PMID 27649880. S2CID 39198636.
- ^ Žumer K, Low AK, Jiang H, Saksela K, Peterlin BM (April 2012). "Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes". Molecular and Cellular Biology. 32 (8): 1354–62. doi:10.1128/mcb.06359-11. PMC 3318594. PMID 22310661.
- ^ Scott HS, Heino M, Peterson P, Mittaz L, Lalioti MD, Betterle C, et al. (August 1998). "Common mutations in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients of different origins". Molecular Endocrinology. 12 (8): 1112–9. doi:10.1210/mend.12.8.0143. PMID 9717837.
- ^ Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, et al. (February 2000). "Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein". American Journal of Human Genetics. 66 (2): 378–92. doi:10.1086/302765. PMC 1288090. PMID 10677297.
- ^ Fardi Golyan F, Ghaemi N, Abbaszadegan MR, Dehghan Manshadi SH, Vakili R, Druley TE, et al. (November 2019). "Novel mutation in AIRE gene with autoimmune polyendocrine syndrome type 1". Immunobiology. 224 (6): 728–733. doi:10.1016/j.imbio.2019.09.004. PMID 31526676. S2CID 202671335.
- ^ "OMIM".
- ^ Ramsey C, Winqvist O, Puhakka L, Halonen M, Moro A, Kämpe O, et al. (February 2002). "Aire deficient mice develop multiple features of APECED phenotype and show altered immune response". Human Molecular Genetics. 11 (4): 397–409. doi:10.1093/hmg/11.4.397. PMID 11854172.
- ^ Iioka T, Furukawa K, Yamaguchi A, Shindo H, Yamashita S, Tsukazaki T (August 2003). "P300/CBP acts as a coactivator to cartilage homeoprotein-1 (Cart1), paired-like homeoprotein, through acetylation of the conserved lysine residue adjacent to the homeodomain". Journal of Bone and Mineral Research. 18 (8): 1419–29. doi:10.1359/jbmr.2003.18.8.1419. PMID 12929931. S2CID 8125330.
Further reading
edit- Björses P, Aaltonen J, Horelli-Kuitunen N, Yaspo ML, Peltonen L (1998). "Gene defect behind APECED: a new clue to autoimmunity". Human Molecular Genetics. 7 (10): 1547–53. doi:10.1093/hmg/7.10.1547. PMID 9735375.
- Heino M, Peterson P, Kudoh J, Shimizu N, Antonarakis SE, Scott HS, Krohn K (September 2001). "APECED mutations in the autoimmune regulator (AIRE) gene". Human Mutation. 18 (3): 205–11. doi:10.1002/humu.1176. PMID 11524731. S2CID 40379449.
- Sato K, Nakajima K, Imamura H, Deguchi T, Horinouchi S, Yamazaki K, et al. (December 2002). "A novel missense mutation of AIRE gene in a patient with autoimmune polyendocrinopathy, candidiasis and ectodermal dystrophy (APECED), accompanied with progressive muscular atrophy: case report and review of the literature in Japan". Endocrine Journal. 49 (6): 625–33. doi:10.1507/endocrj.49.625. PMID 12625412.
- Ruan QG, She JX (March 2004). "Autoimmune polyglandular syndrome type 1 and the autoimmune regulator". Clinics in Laboratory Medicine. 24 (1): 305–17. doi:10.1016/j.cll.2004.01.008. PMID 15157567.
- Holmdahl R (March 2007). "Aire-ing self antigen variability and tolerance". European Journal of Immunology. 37 (3): 598–601. doi:10.1002/eji.200737152. PMID 17323409. S2CID 26685751.
- Aaltonen J, Björses P, Sandkuijl L, Perheentupa J, Peltonen L (September 1994). "An autosomal locus causing autoimmune disease: autoimmune polyglandular disease type I assigned to chromosome 21" (PDF). Nature Genetics. 8 (1): 83–7. doi:10.1038/ng0994-83. hdl:1765/59110. PMID 7987397. S2CID 20365290.
- Aaltonen J, Horelli-Kuitunen N, Fan JB, Björses P, Perheentupa J, Myers R, et al. (August 1997). "High-resolution physical and transcriptional mapping of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy locus on chromosome 21q22.3 by FISH". Genome Research. 7 (8): 820–9. doi:10.1101/gr.7.8.820. PMID 9267805.
- Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. (December 1997). "Positional cloning of the APECED gene". Nature Genetics. 17 (4): 393–8. doi:10.1038/ng1297-393. PMID 9398839. S2CID 1583134.
- Scott HS, Heino M, Peterson P, Mittaz L, Lalioti MD, Betterle C, et al. (August 1998). "Common mutations in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients of different origins". Molecular Endocrinology. 12 (8): 1112–9. doi:10.1210/mend.12.8.0143. PMID 9717837.
- Heino M, Scott HS, Chen Q, Peterson P, Mäebpää U, Papasavvas MP, et al. (1999). "Mutation analyses of North American APS-1 patients". Human Mutation. 13 (1): 69–74. doi:10.1002/(SICI)1098-1004(1999)13:1<69::AID-HUMU8>3.0.CO;2-6. PMID 9888391. S2CID 27558091.
- Björses P, Pelto-Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I (February 1999). "Localization of the APECED protein in distinct nuclear structures". Human Molecular Genetics. 8 (2): 259–66. doi:10.1093/hmg/8.2.259. PMID 9931333.
- Rinderle C, Christensen HM, Schweiger S, Lehrach H, Yaspo ML (February 1999). "AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers". Human Molecular Genetics. 8 (2): 277–90. doi:10.1093/hmg/8.2.277. PMID 9931335.
- Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, et al. (February 2000). "Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein". American Journal of Human Genetics. 66 (2): 378–92. doi:10.1086/302765. PMC 1288090. PMID 10677297.
- Pitkänen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, et al. (June 2000). "The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein". The Journal of Biological Chemistry. 275 (22): 16802–9. doi:10.1074/jbc.M908944199. PMID 10748110.
- Pitkänen J, Vähämurto P, Krohn K, Peterson P (June 2001). "Subcellular localization of the autoimmune regulator protein. characterization of nuclear targeting and transcriptional activation domain". The Journal of Biological Chemistry. 276 (22): 19597–602. doi:10.1074/jbc.M008322200. PMID 11274163.
- Saugier-Veber P, Drouot N, Wolf LM, Kuhn JM, Frébourg T, Lefebvre H (April 2001). "Identification of a novel mutation in the autoimmune regulator (AIRE-1) gene in a French family with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy". European Journal of Endocrinology. 144 (4): 347–51. doi:10.1530/eje.0.1440347. PMID 11275943.
External links
edit- AIRE+protein at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Human AIRE genome location and AIRE gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: O43918 (Autoimmune regulator) at the PDBe-KB.