A nucleolar detention center (DC) is a region of the cell in which certain proteins are temporarily detained in periods of cellular stress.[1] DCs are absent from cells under normal culture conditions, but form in response to specific environmental triggers. The detention of numerous proteins in DCs is believed to reduce metabolic activity and promote survival under unfavorable conditions.[2] DCs form at the center of nucleoli and therefore disrupt the normal organization of these organelles. The structural remodeling that ensues leaves nucleoli unable to sustain their primary function, ribosomal biogenesis. Therefore, the formation of DCs is thought to convert nucleoli from “ribosome factories” to “prisons for proteins”.[1]
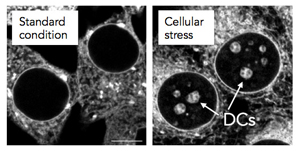
Detention center formation is thought to be controlled by the varying expression of intergenic spacer long noncoding RNA (IGS lncRNA).[1][3] Under normal conditions, the genes that code for IGS lncRNA are silenced. Cellular stressors such as heat shock and acidosis trigger the expression of IGS lncRNA which, in turn, initiates the structural changes that transform the internal domain of the nucleolus into the detention center.[1][4] The actual formation of the detention center domain is facilitated by the binding and sequestration of target proteins by IGS lncRNA. Different types of IGS lncRNA associate selectively with target proteins, temporarily inactivating them and causing them to aggregate in large clumps in the nucleolus.[5] The absence of cellular stressors and return to cellular environment homeostasis decreases IGS lncRNA transcription, causing the nucleolus to relinquish detained proteins, return to its original structural confirmation, and resume the production of ribosomes.[1]
The IGS lncRNA sequences produced in response to cellular stress differ depending on the type of stress-inducing stimulus. IGS lncRNA produced in response to heat shock is transcribed from a different region than IGS lncRNA produced in response to acidosis.[5] The set of proteins sequestered in the detention center is dependent on the type of IGS lncRNA produced, and therefore on the type of environmental stressor present.[5]
References
edit- ^ a b c d e Jacob, Mathieu D.; Audas, Timothy E.; Uniacke, James; Trinkle-Mulcahy, Laura; Lee, Stephen (2013-09-15). "Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus". Molecular Biology of the Cell. 24 (18): 2943–2953. doi:10.1091/mbc.E13-04-0223. ISSN 1939-4586. PMC 3771955. PMID 23904269.
- ^ Audas, Timothy E.; Jacob, Mathieu D.; Lee, Stephen (2012-06-01). "The nucleolar detention pathway: A cellular strategy for regulating molecular networks". Cell Cycle. 11 (11): 2059–2062. doi:10.4161/cc.20140. ISSN 1551-4005. PMC 3368857. PMID 22580471.
- ^ Audas, Timothy E.; Jacob, Mathieu D.; Lee, Stephen (2012-01-27). "Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA". Molecular Cell. 45 (2): 147–157. doi:10.1016/j.molcel.2011.12.012. ISSN 1097-4164. PMID 22284675.
- ^ Prasanth, Kannanganattu V. (2012-01-27). "Policing cells under stress: noncoding RNAs capture proteins in nucleolar detention centers". Molecular Cell. 45 (2): 141–142. doi:10.1016/j.molcel.2012.01.005. ISSN 1097-4164. PMID 22284672.
- ^ a b c Pirogov, Sergei A.; Gvozdev, Vladimir A.; Klenov, Mikhail S. (2019-07-02). "Long Noncoding RNAs and Stress Response in the Nucleolus". Cells. 8 (7): 668. doi:10.3390/cells8070668. ISSN 2073-4409. PMC 6678565. PMID 31269716.