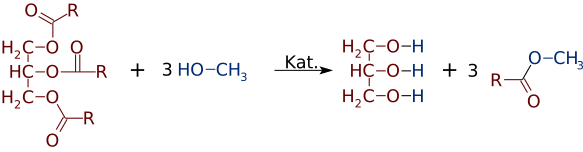Fatty acid methyl esters (FAME) are a type of fatty acid ester that are derived by transesterification of fats with methanol. The molecules in biodiesel are primarily FAME, usually obtained from vegetable oils by transesterification. They are used to produce detergents and biodiesel.[1] FAME are typically produced by an alkali-catalyzed reaction between fats and methanol in the presence of base such as sodium hydroxide, sodium methoxide[2] or potassium hydroxide. One reason for using FAME (fatty acid methyl esters) in biodiesel production, rather than free fatty acids, is to mitigate the potential corrosion they can cause to metals of engines, production facilities, and related infrastructure. While free fatty acids are only mildly acidic, over time they can lead to cumulative corrosion. In contrast, their esters, such as FAME, are less corrosive and therefore preferred for biodiesel production. As an improved quality, FAMEs also usually have about 12-15 units higher cetane number than their unesterified counterparts.[3]
Selected FAMEs
editFAMEs are colorless compounds with melting points near room temperature.
| FAME | formula | Registry number | refractive index | density (g/cm3) | melting point (°C) | boiling point (°C) |
|---|---|---|---|---|---|---|
| Methyl laurate | CH3(CH2)10CO2CH3 | 111-82-0 | 1.4301 | 0.8702 | 5.2 | 267 |
| Methyl myristate | CH3(CH2)12CO2CH3 | 124-10-7 | - | 0.8671 | 17.8 | 295 |
| Methyl palmitate | CH3(CH2)14CO2CH3 | 112-39-0 | 1.4310 | - | 29.5 | 417 |
| Methyl stearate | CH3(CH2)16CO2CH3 | 112-61-8 | 1.45740 | 0.83 | 40 | 443 |
| Methyl oleate | CH3(CH2)6CH=CH(CH2)5CO2CH3 | 112-62-9 | 112- | 0.8739 | -19.9 | - |
FAME profiling
editMicroorganisms have diverse and sometimes distinctive FAME profiles, the basis of "microbial fingerprinting". After triglycerides, fatty acids and certain other lipids of some cultured microbes are transesterified or esterified, the resulting FAMEs can be analyzed with gas chromatography.[4] These profiles can be used as a tool for microbial source tracking (MST) to identify pathological bacteria strains[5] and for characterizing new species of bacteria.
For example, a profile created from cultured bacteria from some water sample can be compared to a profile of known pathological bacteria to find out if the water is polluted by feces or not.[5]
Unusual FAMEs
editIn June 2022, Polycyclopropanated fatty acid methyl ester (POP-FAME) fuels were biosynthesized from Streptomyces coelicolor bacteria, which have energy densities of more than 50MJ/L larger than the most widely used aviation and rocket fuels.[6]
See also
editReferences
edit- ^ Anneken, David J.; Both, Sabine; Christoph, Ralf; Fieg, Georg; Steinberner, Udo; Westfechtel, Alfred (2006). "Fatty Acids". Ullmann's Encyclopedia of Industrial Chemistry. Weinheim: Wiley-VCH. doi:10.1002/14356007.a10_245.pub2. ISBN 9783527306732. OCLC 910197915.
- ^ Vyas, Amish P.; Verma, Jaswant L.; Subrahmanyam, N. (2010). "A review on FAME production processes". Fuel. 89 (1): 1–9. doi:10.1016/j.fuel.2009.08.014. ISSN 0016-2361.
- ^ Schobert, Harold H. (2013). Chemistry of Fossil Fuels and Biofuels. Cambridge, NY: Cambridge University Press. pp. 62–64. doi:10.1017/CBO9780511844188. ISBN 9780511844188. OCLC 823724682.
- ^ Sekora, Nicholas S.; Lawrence, Kathy S.; Agudelo, Paula; van Santen, Edzard; McInroy, John A. (2009). "Using FAME Analysis to Compare, Differentiate, and Identify Multiple Nematode Species". Journal of Nematology. 41 (3): 163–173. PMC 3380492. PMID 22736811.
- ^ a b Duran, Metin; Haznedaroğlu, Berat Z.; Zitomer, Daniel H. (2006). "Microbial source tracking using host specific FAME profiles of fecal coliforms". Water Research. 40 (1): 67–74. doi:10.1016/j.watres.2005.10.019. PMID 16360192.
- ^ Cruz-Morales, Pablo; Yin, Kevin; Landera, Alexander; Cort, John R.; Young, Robert P.; Kyle, Jennifer E.; Bertrand, Robert; Iavarone, Anthony T.; Acharya, Suneil; Cowan, Aidan; Chen, Yan; Gin, Jennifer W.; Scown, Corinne D.; Petzold, Christopher J.; Araujo-Barcelos, Carolina (2022-07-20). "Biosynthesis of polycyclopropanated high energy biofuels". Joule. 6 (7): 1590–1605. doi:10.1016/j.joule.2022.05.011. ISSN 2542-4785. S2CID 250189786.