Protoporphyrin ferrochelatase (EC 4.98.1.1, formerly EC 4.99.1.1, or ferrochelatase; systematic name protoheme ferro-lyase (protoporphyrin-forming)) is an enzyme encoded by the FECH gene in humans.[1] Ferrochelatase catalyses the eighth and terminal step in the biosynthesis of heme, converting protoporphyrin IX into heme B. It catalyses the reaction:
| Protoporphyrin ferrochelatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Ferrochelatase homodimer, Human | |||||||||
| Identifiers | |||||||||
| EC no. | 4.98.1.1 | ||||||||
| CAS no. | 9012-93-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Ferrochelatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Human ferrochelatase | |||||||||
| Identifiers | |||||||||
| Symbol | Ferrochelatase | ||||||||
| Pfam | PF00762 | ||||||||
| InterPro | IPR001015 | ||||||||
| PROSITE | PDOC00462 | ||||||||
| SCOP2 | 1ak1 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 129 | ||||||||
| OPM protein | 1hrk | ||||||||
| |||||||||
- protoporphyrin + Fe2+ → protoheme + 2 H+
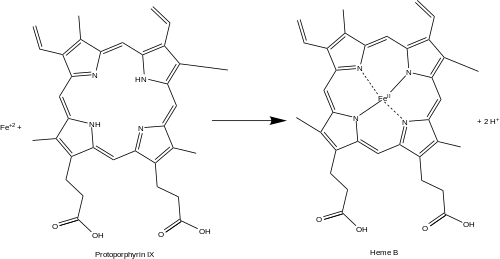
Function
editFerrochelatase catalyzes the insertion of ferrous iron into protoporphyrin IX in the heme biosynthesis pathway to form heme B. The enzyme is localized to the matrix-facing side of the inner mitochondrial membrane. Ferrochelatase is the best known member of a family of enzymes that add divalent metal cations to tetrapyrrole structures.[2] For example, magnesium chelatase adds magnesium to protoporphyrin IX in the first step of bacteriochlorophyll biosynthesis.[3]
Heme B is an essential cofactor in many proteins and enzymes. In particular, heme b plays a key role as the oxygen carrier in hemoglobin in red blood cells and myoglobin in muscle cells. Furthermore, heme B is found in cytochrome b, a key component in Q-cytochrome c oxidoreductase (complex III) in oxidative phosphorylation.[4]
Structure
editHuman ferrochelatase is a homodimer composed of two 359 amino acid polypeptide chains. It has a total molecular weight of 85.07 kDa.[5] Each subunit is composed of five regions: a mitochondrial localization sequence, the N terminal domain, two folded domains, and a C terminal extension. Residues 1–62 form a mitochondrial localization domain that is cleaved in post-translational modification. The folded domains contain a total of 17 α-helices and 8 β-sheets. The C terminal extension contains three of the four cysteine residues (Cys403, Cys406, Cys411) that coordinate the catalytic iron–sulfur cluster (2Fe-2S). The fourth coordinating cysteine resides in the N-terminal domain (Cys196).[6]
The active pocket of ferrocheltase consists of two hydrophobic "lips" and a hydrophilic interior. The hydrophobic lips, consisting of the highly conserved residues 300–311, face the inner mitochondrial membrane and facilitate the passage of the poorly soluble protoporphyrin IX substrate and the heme product via the membrane. The interior of the active site pocket contains a highly conserved acidic surface that facilitates proton extraction from protoporphyrin. Histidine and aspartate residues roughly 20 angstroms from the center of the active site on the mitochondrial matrix side of the enzyme coordinate metal binding.[6]
Mechanism
editThe mechanism of human protoporphyrin metalation remains under investigation. Many researchers have hypothesized distortion of the porphyrin macrocycle as key to catalysis. Researchers studying Bacillus subtilis ferrochelatase propose a mechanism for iron insertion into protoporphyrin in which the enzyme tightly grips rings B, C, and D while bending ring A 36o. Normally planar, this distortion exposes the lone pair of electrons on the nitrogen in ring A to the Fe+2 ion.[2] Subsequent investigation revealed a 100o distortion in protoporphyrin bound to human ferrochelatase. A highly conserved histidine residue (His183 in B. subtilis, His263 in humans) is essential for determining the type of distortion, as well as acting as the initial proton acceptor from protoporphyrin.[6][7] Anionic residues form a pathway facilitating proton movement away from the catalytic histidine.[6] Frataxin chaperones iron to the matrix side of ferrochelatase, where aspartate and histidine residues on both proteins coordinate iron transfer into ferrochelatase.[8] Two arginine and tyrosine residues in the active site (Arg164, Tyr165) may perform the final metalation.[6]
Clinical significance
editDefects in ferrochelatase create a buildup of protoporphyrin IX, causing erythropoietic protoporphyria (EPP).[9] The disease can result from a variety of mutations in FECH, most of which behave in an autosomal dominant manner with low clinical penetrance. Clinically, patients with EPP present with a range of symptoms, from asymptomatic to suffering from an extremely painful photosensitivity. In less than five percent of cases, accumulation of protoporphyrin in the liver results in cholestasis (blockage of bile flow from the liver to the small intestine) and terminal liver failure.[10]
In cases of lead poisoning, lead inhibits ferrochelatase activity, in part resulting in porphyria.[11]
Interactions
editFerrochelatase interacts with numerous other enzymes involved in heme biosynthesis, catabolism, and transport, including protoporphyrinogen oxidase, 5-aminolevulinate synthase, ABCB10, ABCB7, succinyl-CoA synthetase,[12] and mitoferrin-1.[13] Multiple studies have suggested the existence of an oligomeric complex that enables substrate channeling and coordination of overall iron and porphyrin metabolism throughout the cell.[12][13] N-methylmesoporphyrin (N-MeMP) is a competitive inhibitor with protoporphyrin IX and is thought to be a transition state analog. As such, N-MeMP has been used extensively as a stabilizing ligand for x-ray crystallography structure determination.[14] Frataxin acts as the Fe+2 chaperone and complexes with ferrochelatase on its mitochondrial matrix side.[8] Ferrochelatase can also insert other divalent metal ions into protoporphyrin. Some ions, such as Zn+2, Ni, and Co form other metalloporphyrins while heavier metal ions such as Mn, Pb, Hg, and Cd inhibit product release after metallation.[15]
See also
editReferences
edit- ^ "FECH - Ferrochelatase, mitochondrial precursor - Homo sapiens (Human) - FECH gene & protein".
- ^ a b Lecerof, D.; Fodje, M.; Hansson, A.; Hansson, M.; Al-Karadaghi, S. (March 2000). "Structural and mechanistic basis of porphyrin metallation by ferrochelatase". Journal of Molecular Biology. 297 (1): 221–232. doi:10.1006/jmbi.2000.3569. PMID 10704318.
- ^ Leeper, F. J. (1985). "The biosynthesis of porphyrins, chlorophylls, and vitamin B12". Natural Product Reports. 2 (1): 19–47. doi:10.1039/NP9850200019. PMID 3895052.
- ^ Berg, Jeremy; Tymoczko, John; Stryer, Lubert (2012). Biochemistry (7th ed.). New York: W.H. Freeman. ISBN 9781429229364.
- ^ "RCSB PDB - 1Hrk: Crystal Structure of Human Ferrochelatase".
- ^ a b c d e Wu, Chia-Kuei; Dailey, Harry A.; Rose, John P.; Burden, Amy; Sellers, Vera M.; Wang, Bi-Cheng (1 February 2001). "The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis". Nature Structural Biology. 8 (2): 156–160. doi:10.1038/84152. PMID 11175906. S2CID 9822420.
- ^ Karlberg, Tobias; Hansson, Mattias D.; Yengo, Raymond K.; Johansson, Renzo; Thorvaldsen, Hege O.; Ferreira, Gloria C.; Hansson, Mats; Al-Karadaghi, Salam (May 2008). "Porphyrin Binding and Distortion and Substrate Specificity in the Ferrochelatase Reaction: The Role of Active Site Residues". Journal of Molecular Biology. 378 (5): 1074–1083. doi:10.1016/j.jmb.2008.03.040. PMC 2852141. PMID 18423489.
- ^ a b Bencze, Krisztina Z.; Yoon, Taejin; Mill?n-Pacheco, C?sar; Bradley, Patrick B.; Pastor, Nina; Cowan, J. A.; Stemmler, Timothy L. (2007). "Human frataxin: iron and ferrochelatase binding surface". Chemical Communications (18): 1798–1800. doi:10.1039/B703195E. PMC 2862461. PMID 17476391.
- ^ James, William D.; Berger, Timothy G. (2006). Andrews' Diseases of the Skin: clinical Dermatology. Saunders Elsevier. ISBN 0-7216-2921-0.
- ^ Rüfenacht, U.B.; Gouya, L.; Schneider-Yin, X.; Puy, H.; Schäfer, B.W.; Aquaron, R.; Nordmann, Y.; Minder, E.I.; Deybach, J.C. (1998). "Systematic Analysis of Molecular Defects in the Ferrochelatase Gene from Patients with Erythropoietic Protoporphyria". The American Journal of Human Genetics. 62 (6): 1341–52. doi:10.1086/301870. PMC 1377149. PMID 9585598.
- ^ "Lead Toxicity -- What Are Possible Health Effects from Lead Exposure?". Agency for Toxic Substances & Disease Registry. Retrieved 9 February 2021.
- ^ a b Medlock, Amy E.; Shiferaw, Mesafint T.; Marcero, Jason R.; Vashisht, Ajay A.; Wohlschlegel, James A.; Phillips, John D.; Dailey, Harry A.; Liesa, Marc (19 August 2015). "Identification of the Mitochondrial Heme Metabolism Complex". PLOS ONE. 10 (8): e0135896. Bibcode:2015PLoSO..1035896M. doi:10.1371/journal.pone.0135896. PMC 4545792. PMID 26287972.
- ^ a b Chen, W.; Dailey, H. A.; Paw, B. H. (28 April 2010). "Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis". Blood. 116 (4): 628–630. doi:10.1182/blood-2009-12-259614. PMC 3324294. PMID 20427704.
- ^ Medlock, A.; Swartz, L.; Dailey, T. A.; Dailey, H. A.; Lanzilotta, W. N. (29 January 2007). "Substrate interactions with human ferrochelatase". Proceedings of the National Academy of Sciences. 104 (6): 1789–1793. Bibcode:2007PNAS..104.1789M. doi:10.1073/pnas.0606144104. PMC 1794275. PMID 17261801.
- ^ Medlock, Amy E.; Carter, Michael; Dailey, Tamara A.; Dailey, Harry A.; Lanzilotta, William N. (October 2009). "Product Release Rather than Chelation Determines Metal Specificity for Ferrochelatase". Journal of Molecular Biology. 393 (2): 308–319. doi:10.1016/j.jmb.2009.08.042. PMC 2771925. PMID 19703464.
Further reading
edit- Cox TM (June 1997). "Erythropoietic protoporphyria". Journal of Inherited Metabolic Disease. 20 (2): 258–69. doi:10.1023/A:1005317124985. PMID 9211198. S2CID 12493042.
- Brenner DA, Didier JM, Frasier F, Christensen SR, Evans GA, Dailey HA (June 1992). "A molecular defect in human protoporphyria". American Journal of Human Genetics. 50 (6): 1203–10. PMC 1682545. PMID 1376018.
- Nakahashi Y, Fujita H, Taketani S, Ishida N, Kappas A, Sassa S (January 1992). "The molecular defect of ferrochelatase in a patient with erythropoietic protoporphyria". Proceedings of the National Academy of Sciences of the United States of America. 89 (1): 281–5. Bibcode:1992PNAS...89..281N. doi:10.1073/pnas.89.1.281. PMC 48220. PMID 1729699.
- Lamoril J, Boulechfar S, de Verneuil H, Grandchamp B, Nordmann Y, Deybach JC (December 1991). "Human erythropoietic protoporphyria: two point mutations in the ferrochelatase gene". Biochemical and Biophysical Research Communications. 181 (2): 594–9. doi:10.1016/0006-291X(91)91231-Z. PMID 1755842.
- Nakahashi Y, Taketani S, Okuda M, Inoue K, Tokunaga R (December 1990). "Molecular cloning and sequence analysis of cDNA encoding human ferrochelatase". Biochemical and Biophysical Research Communications. 173 (2): 748–55. doi:10.1016/S0006-291X(05)80099-3. PMID 2260980.
- Rossi E, Attwood PV, Garcia-Webb P, Costin KA (May 1990). "Inhibition of human lymphocyte ferrochelatase activity by hemin". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1038 (3): 375–81. doi:10.1016/0167-4838(90)90251-A. PMID 2340297.
- Polson RJ, Lim CK, Rolles K, Calne RY, Williams R (September 1988). "The effect of liver transplantation in a 13-year-old boy with erythropoietic protoporphyria". Transplantation. 46 (3): 386–9. doi:10.1097/00007890-198809000-00010. PMID 3047929.
- Bonkovsky HL, Schned AR (January 1986). "Fatal liver failure in protoporphyria. Synergism between ethanol excess and the genetic defect". Gastroenterology. 90 (1): 191–201. doi:10.1016/0016-5085(86)90093-4. PMID 3940245.
- Prasad AR, Dailey HA (August 1995). "Effect of cellular location on the function of ferrochelatase". The Journal of Biological Chemistry. 270 (31): 18198–200. doi:10.1074/jbc.270.31.18198. PMID 7629135.
- Sarkany RP, Alexander GJ, Cox TM (June 1994). "Recessive inheritance of erythropoietic protoporphyria with liver failure". Lancet. 343 (8910): 1394–6. doi:10.1016/S0140-6736(94)92525-9. PMID 7910885. S2CID 42243172.
- Tugores A, Magness ST, Brenner DA (December 1994). "A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene". The Journal of Biological Chemistry. 269 (49): 30789–97. doi:10.1016/S0021-9258(18)47351-6. PMID 7983009.
- Dailey HA, Sellers VM, Dailey TA (January 1994). "Mammalian ferrochelatase. Expression and characterization of normal and two human protoporphyric ferrochelatases". The Journal of Biological Chemistry. 269 (1): 390–5. doi:10.1016/S0021-9258(17)42362-3. PMID 8276824.
- Wang X, Poh-Fitzpatrick M, Carriero D, Ostasiewicz L, Chen T, Taketani S, Piomelli S (April 1993). "A novel mutation in erythropoietic protoporphyria: an aberrant ferrochelatase mRNA caused by exon skipping during RNA splicing". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1181 (2): 198–200. doi:10.1016/0925-4439(93)90112-e. PMID 8481408.
- Nakahashi Y, Miyazaki H, Kadota Y, Naitoh Y, Inoue K, Yamamoto M, Hayashi N, Taketani S (May 1993). "Molecular defect in human erythropoietic protoporphyria with fatal liver failure". Human Genetics. 91 (4): 303–6. doi:10.1007/BF00217346. PMID 8500787. S2CID 5844599.
- Imoto S, Tanizawa Y, Sato Y, Kaku K, Oka Y (July 1996). "A novel mutation in the ferrochelatase gene associated with erythropoietic protoporphyria". British Journal of Haematology. 94 (1): 191–7. doi:10.1046/j.1365-2141.1996.d01-1771.x. PMID 8757534. S2CID 27290533.
- Crouse BR, Sellers VM, Finnegan MG, Dailey HA, Johnson MK (December 1996). "Site-directed mutagenesis and spectroscopic characterization of human ferrochelatase: identification of residues coordinating the [2Fe-2S] cluster". Biochemistry. 35 (50): 16222–9. doi:10.1021/bi9620114. PMID 8973195.
External links
edit- UMich Orientation of Proteins in Membranes protein/pdbid-1hrk
- Ferrochelatase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)