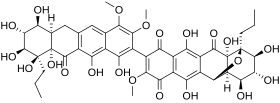
Hibarimicinone is an organic atropisomeric small molecule, derived from hibarimicins, which are isolated from the microbial culture strain of Microbispora rosea from the Hibari region of Japan.[2][3][4][5] Analysis of the bacteria identified a new class of molecule containing a dimeric-tetracyclic polyketide backbone, which are now known as the hibarimicins. Hibarimicinone and its derivatives were initially extracted for their potential inhibitory properties of various protein kinases, such as calmodulin-dependent protein kinase III (CAMKIII), protein kinase A (PKA), protein kinase C (PKC), and protein tyrosine kinase (PTK).[3] The atropisomerism that the hibarimicin family possess arises from the hindered rotation about the biaryl axis connecting to the two monomers, with steric hindrance imposed by the four ortho substituents.[6] The absolute configuration around the axis was elucidated by Romaine et al., utilizing a C2-symmetric shunt metabolite HMP-Y6, a glycosylated structural analogue of hibarimicinone, in tandem with biomimetic homocoupling.[7] Recent work by multiple groups have shown the total synthesis of hibarimicinone and its derivatives.[8][9][10]
| |
| Names | |
|---|---|
| Preferred IUPAC name
(2R,5S,6R,7S,8S,9S)-15-[(6aR,7S,8R,9S,10S,10aS)-1,7,8,9,10,10a,12-Heptahydroxy-3,4-dimethoxy-11-oxo-10-propyl-6,6a,7,8,9,10,10a,11-octahydro-2-tetracenyl]-5,6,7,9,12,19-hexahydroxy-16-methoxy-4-propyl ;-3-oxapentacyclo[9.8.0.02,8.04,9.013,18]nonadeca-1(11),12,15,18-tetraene-10,14,17-trione | |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C45H48O21 | |
| Molar mass | 924.858 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Biosynthesis
editThe biosynthesis of hibarimicinone was initially hypothesized to be produced from a polyketide synthase. Through 13C-labelling and blocked mutants of the TP-A0121 strain, Oki et al. demonstrated that hibarimicinone is produced by an oxidative coupling of two tetracyclic polyketides, which undergo an oxidative cyclization to generate the ether ring.[3][5][7] The hibarimicinone monomer is formed initially by a Type II polyketide synthase. The 22 carbon chain is initiated by butyryl-CoA and its decarboxylative addition to a malonyl-CoA derived two carbon ketide unit. The KS/CLF binds to the starting butyryl-CoA, freeing the ACP domain to bind to another malonate group from malonyl-CoA. Thioester exchange occurs between the KS/CLF and the ACP; the cycle then repeats nine more times to produce the full carbon chain (1).[11] Multiple aldol additions occur to form four six-membered cyclization and aromization (2). The CLF/KS subunit is cleaved off by a thioesterase and modified by numerous steps (four oxidations, two NADPH dependent reductions, two methylations and decarboxylation) to form the final monomeric unit (3). Two monomer units undergo oxidative coupling to form the atropisomeric axis (4). Oxidative cyclization occurs on one end of the molecule to form the ether ring (5). Finally, the hydroquinone on the same side of the previously mentioned ether oxidizes to form the quinone species, giving hibarimicinone.
References
edit- ^ "KNApSAcK Metabolite Information - C00015176". www.knapsackfamily.com.
- ^ Hori, H.; Higashi, K.; Ishiyama, T.; Uramoto, M.; Uehara, Y.; Oki, T. Tetrahedron Lett. 1996 37, 2785-2788
- ^ a b c Kajiura, T.; Furumai, T.; Igarashi, Y.; Hori, H.; Higashi, K.; Ishiyama, T.; Uramoto, M.; Uehara, Y.; Oki, T. J. Antibiot. 1998, 51, 394-401
- ^ Hori, H.; Kajiura, T.; Igarashi, Y.; Hori, H.; Higashi, K; Ishiyama, T.; Uramoto, M; Uehara, Y.; Oki, T. J. Antibiot. 2002, 55, 53-60
- ^ a b Hori, H.; Kajiura, T.; Igarashi, Y.; Furumai, T; Higashi, K.; Ishiyama, T; Uramoto, M; Uehara, Y; Oki, T. J. Antibiot., 2002, 55, 46-52
- ^ Smyth, J.; Butler, N.; Keller, P. Nat. Prod. Rep. 2015, 32, 1562-1583
- ^ a b Romaine, I. M.; Hempel, J. E.; Shanmugam, G.; Hori, H.; Igarashi, Y.; Polavarpu, P. L.; Sulikowshi, G. A. Org. Lett. 2011, 13, 4538-4541
- ^ B. B. Liau, B. C. Milgram and M. D. Shair, J. Am. Chem. Soc., 2012, 134, 16765-16772
- ^ Tatsuta, K.; Fukuda, T.; Ishimori, T.; Yachi, R.; Yoshida, S.; Hashimoto, H.; Hosokawa, S. Tetrahedron Lett., 2012, 53, 422-425
- ^ Tatsuta, K.; Hosokawa, S. Chem. Rec., 2014, 14, 28-40
- ^ Dewick, Paul M. (2002). Medicinal Natural Products. John Wiley and Sons, LTD