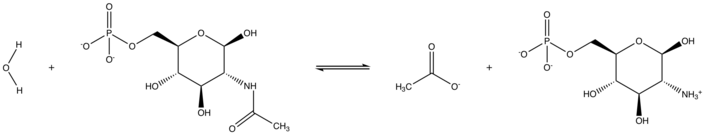
In enzymology, N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25), also known as GlcNAc-6-phosphate deacetylase or NagA, is an enzyme that catalyzes the deacetylation of N-acetylglucosamine-6-phosphate (GlcNAc-6-P) to glucosamine-6-phosphate (GlcN-6-P):
| N-acetylglucosamine-6-phosphate deacetylase in Mycobacterium smegmatis | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 3.5.1.25 | ||||||||
| CAS no. | 9027-50-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
- H2O + N-acetyl-D-glucosamine 6-phosphate acetate + D-glucosamine 6-phosphate[1]
NagA Reaction
GlcNAc-6-phosphate deacetylase is encoded by the gene NagA.[2]
This enzyme belongs to the amidohydrolase superfamily.[3] Amidohydrolases are a type of hydrolase that acts upon amide bonds. All members of the amidohydrolase family employ a TIM barrel structure, and a vast majority of members are metalloenzymes.[4] The family of enzymes is important in amino acid and nucleotide metabolism as well as biodegradation of agricultural and industrial compounds. NagA participates in amino-sugar metabolism, specifically in the biosynthesis of amino-sugar-nucleotides.[5]
Structure
editNagA is a homodimeric enzyme with two domains in each dimer of the structure.[6] Each domain I comprises a (β/α)8 - barrel structural fold, also known as a TIM barrel, and contains an active site of the enzyme. Each active site consists of the catalytic site of the enzyme and the metal-binding site that are involved in substrate and metal co-factor recognition, respectively. Domain I also forms the dimeric interface with domain I of the neighboring subunit.[6] The smaller second domain of NagA enzymes comprises a β-barrel, which potentially acts to stabilize the enzyme.[6] While all members of the amidohydrolase superfamily employ a TIM-barrel structural fold, NagA in Escherichia coli (EcNagA) has a pseudo-TIM barrel enclosing the funnel-like catalytic site of the enzyme.[7] The dimer structure of NagA is considered crucial for the activity and thermostability of the enzyme.[8]
Metal-binding site
editAmidohydrolase enzymes can bind one, two, or three metal atoms in the active site. These metals can include Zn2+, Co2+, Fe2+, Cd2+, and others.[1] EcNagA contains a mononuclear metal-binding site with a Zn2+ ion;[3] in addition, EcNagA shows a phosphate ion bound at the metal-binding site.[7] Unlike EcNagA, NagA of Mycobacterium smegmatis (MSNagA) and Bacillus subtilis (BsNagA) have binuclear metal-binding sites. MSNagA has two divalent metal ions located in each active site, which are both required for efficient catalysis and structural stability.[6] While most other bacteria species use Zn as their metal co-factor, BsNagA utilizes iron as the predominant metal in the metal-binding site.[9]
Catalytic-binding site
editMost of the active site residues of EcNagA and BsNagA are conserved and share similar structural positions. A notable difference between mycobacterial NagA enzymes and NagA enzymes from other bacterial species is the presence of a cysteine at position 131. Other bacterial species have a lysine residue at this position. This cysteine is located in the flexible loop, which prevents the physiological substrate from binding.[6]
Mechanism
editThe catalytic mechanism for NagA enzymes proposed utilizes nucleophilic attack via a metal-coordinated water molecule or hydroxide ion. The mechanism proceeds via a strictly conserved active-site aspartic acid residue (Asp-273) that acts initially as a base to activate the hydrolytic water molecule in order to attack the carbonyl group of the substrate.[3] Asp-273 then acts as an acid to protonate the amine leaving group. One proposed mechanism using the BsNagA and its two iron co-factors in the metal-binding site demonstrates the nucleophilic attack by an Fe-bridged hydroxide and then the stabilization of the carbonyl oxygen by one of the two Fe atoms.[9]
Biological Function
editNagA is located in the cytoplasm of the cell. N-acetylglucosamine (GlcNAc) enters the cell as part of the breakdown of the cell wall. GlcNAc, a monosaccharide and derivative of glucose, is part of a biopolymer in the bacterial cell wall. This biopolymer forms a layered structure called peptidoglycan (PG). GlcNAc is then converted into GlcNAc-6-P by the enzyme NagE.[10] This substrate is then deacetylated into acetate and GlcN-6-P by NagA.[11] NagA is important for the production of GlcN-6-P, which is then used in two main pathways: PG recycling pathway and the glycolysis pathway.
PG recycling pathway
editIn the PG Recycling pathway, once GlcNAc-6-P is metabolized by NagA, its product, GlcN-6-P, can then be converted to GlcN-1-P by the enzyme GlmM, followed by reacetylation and reaction with UTP by GlmU to form UDP-GlcNAc.[10][11] UDP-GlcNAc is the end product of this pathway, which is then used to make glycosaminoglycans, proteoglycans, and glycolipids, which are all necessary in order to replenish PG for the cell wall.[12] PG recycling is necessary for bacterial cells in order to ensure bacteria growth and prevent cell lysis.[13]
Glycolysis pathway
editInstead of entering the PG recycling pathway, GlcN-6-P can be converted into fructose-6-phosphate by NagB. This reaction is reversible by the enzyme GlmS,[10][11] an amidotransferase.[13] The produced fructose-6-phosphate then enters the glycolysis pathway. Glycolysis catalyzes the production of pyruvate, leading to the citric acid cycle and allowing for the production of amino acids.[14] GlcN-6-P and fructose-6-phosphate act as allosteric regulators of NagA, inhibiting further deacetylation of GlcNAc-6-P.[15]
Disease relevance
editNagA is a potential drug target of Mycobacterium tuberculosis (Mtb). Eliminating NagA produces high levels of the allosteric activator GlcNAc-6-P,[2] which prevents the production of GlcN-6-P in order to proceed with the PG recycling pathway. NagA is, therefore, at a crucial metabolic chokepoint in Mtb,[16] representing the key enzymatic step in the generation of essential amino-sugar precursors. These precursors are required for Mtb cell wall biosynthesis and influence the PG recycling pathway. Additionally, the presence of cysteine in MSNagA's active site may represent a unique exploitative target in Mtb therapeutics.[6]
Structural studies
editAs of early 2019, 11 structures have been solved for this class of enzymes, with PDB accession codes 1O12, 1UN7, 1YMY, 1YRR, 2P50, 2P53, 6FV3, 6FV4, 3EGJ, 3IV8, and 2VHL.
Nomenclature
editThe systematic name of this enzyme class is N-acetyl-D-glucosamine-6-phosphate amidohydrolase. Other names in common use include acetylglucosamine phosphate deacetylase, acetylaminodeoxyglucosephosphate acetylhydrolase, and 2-acetamido-2-deoxy-D-glucose-6-phosphate amidohydrolase.[15]
References
edit- ^ a b "nagA - N-acetylglucosamine-6-phosphate deacetylase - Escherichia coli (strain K12) - nagA gene & protein". www.uniprot.org. Retrieved 2019-03-14.
- ^ a b Alvarez-Añorve LI, Bustos-Jaimes I, Calcagno ML, Plumbridge J (October 2009). "Allosteric regulation of glucosamine-6-phosphate deaminase (NagB) and growth of Escherichia coli on glucosamine". Journal of Bacteriology. 191 (20): 6401–7. doi:10.1128/JB.00633-09. PMC 2753035. PMID 19700525.
- ^ a b c Hall RS, Xiang DF, Xu C, Raushel FM (July 2007). "N-Acetyl-D-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion". Biochemistry. 46 (27): 7942–52. doi:10.1021/bi700543x. PMC 2533526. PMID 17567047.
- ^ Liu A, Huo L (2014-08-15), John Wiley & Sons Ltd (ed.), "Amidohydrolase Superfamily", eLS, John Wiley & Sons, Ltd, doi:10.1002/9780470015902.a0020546.pub2, ISBN 9780470015902
- ^ Yadav V, Panilaitis B, Shi H, Numuta K, Lee K, Kaplan DL (2011-06-02). "N-acetylglucosamine 6-phosphate deacetylase (nagA) is required for N-acetyl glucosamine assimilation in Gluconacetobacter xylinus". PLOS ONE. 6 (6): e18099. Bibcode:2011PLoSO...618099Y. doi:10.1371/journal.pone.0018099. PMC 3107205. PMID 21655093.
- ^ a b c d e f Ahangar MS, Furze CM, Guy CS, Cooper C, Maskew KS, Graham B, Cameron AD, Fullam E (June 2018). "Mycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA)". The Journal of Biological Chemistry. 293 (25): 9770–9783. doi:10.1074/jbc.RA118.002597. PMC 6016474. PMID 29728457.
- ^ a b Ferreira FM, Mendoza-Hernandez G, Castañeda-Bueno M, Aparicio R, Fischer H, Calcagno ML, Oliva G (June 2006). "Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli". Journal of Molecular Biology. 359 (2): 308–21. doi:10.1016/j.jmb.2006.03.024. PMID 16630633.
- ^ Mine S, Kado Y, Watanabe M, Fukuda Y, Abe Y, Ueda T, Kawarabayasi Y, Inoue T, Ishikawa K (November 2014). "The structure of hyperthermophilic β-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site". The FEBS Journal. 281 (22): 5092–103. doi:10.1111/febs.13049. PMID 25227262. S2CID 21178562.
- ^ a b Vincent F, Yates D, Garman E, Davies GJ, Brannigan JA (January 2004). "The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily". The Journal of Biological Chemistry. 279 (4): 2809–16. doi:10.1074/jbc.M310165200. PMID 14557261.
- ^ a b c Park JT, Uehara T (June 2008). "How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan)". Microbiology and Molecular Biology Reviews. 72 (2): 211–27, table of contents. doi:10.1128/MMBR.00027-07. PMC 2415748. PMID 18535144.
- ^ a b c Plumbridge J (September 2009). "An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli". Journal of Bacteriology. 191 (18): 5641–7. doi:10.1128/JB.00448-09. PMC 2737974. PMID 19617367.
- ^ Milewski S, Gabriel I, Olchowy J (January 2006). "Enzymes of UDP-GlcNAc biosynthesis in yeast". Yeast. 23 (1): 1–14. doi:10.1002/yea.1337. PMID 16408321. S2CID 39940329.
- ^ a b Dhar S, Kumari H, Balasubramanian D, Mathee K (January 2018). "Cell-wall recycling and synthesis in Escherichia coli and Pseudomonas aeruginosa - their role in the development of resistance". Journal of Medical Microbiology. 67 (1): 1–21. doi:10.1099/jmm.0.000636. PMID 29185941.
- ^ Stryer L, Tymoczko JL, Berg JM (2002). "The Citric Acid Cycle". Biochemistry. 5th Edition.
- ^ a b White RJ, Pasternak CA (October 1967). "The purification and properties of N-acetylglucosamine 6-phosphate deacetylase from Escherichia coli". The Biochemical Journal. 105 (1): 121–5. doi:10.1042/bj1050121. PMC 1198282. PMID 4861885.
- ^ "Molecular insights of NagA enzyme could help combat TB". News-Medical.net. 2018-07-12. Retrieved 2019-03-11.
Further reading
edit- Yamano N, Matsushita Y, Kamada Y, Fujishima S, Arita M (August 1996). "Purification and characterization of N-acetylglucosamine 6-phosphate deacetylase with activity against N-acetylglucosamine from Vibrio cholerae non-O1". Bioscience, Biotechnology, and Biochemistry. 60 (8): 1320–3. doi:10.1271/bbb.60.1320. PMID 8987551.

