Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA's, there is a disturbance caused in translation.[1] Ribosomal pausing occurs in both eukaryotes and prokaryotes.[2][3] A more severe pause is known as a ribosomal stall.[4]
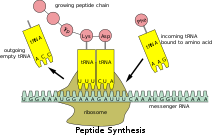
It's been known since the 1980s that different mRNAs are translated at different rates. The main reason for these differences was thought to be the concentration of varieties of rare tRNAs limiting the rate at which some transcripts could be decoded.[5] However, with research techniques such as ribosome profiling, it was found that at certain sites there were higher concentrations of ribosomes than average, and these pause sites were tested with specific codons. No link was found between the occupancy of specific codons and amount of their tRNAs. Thus, the early findings about rare tRNAs causing pause sites don't seem plausible.[2]
Two techniques can localize the ribosomal pause site in vivo; a micrococcal nuclease protection assay and isolation of polysomal transcript.[6] Isolation of polysomal transcripts occurs by centrifuging tissue extracts through a sucrose cushion with translation elongation inhibitors, for example cycloheximide.[7]
Ribosome pausing can be detected during preprolactin synthesis on free polysomes, when the ribosome is paused the other ribosomes are tightly stacked together. When the ribosome pauses, during translation, the fragments that started to translate before the pause took place are overrepresented. However, along with the mRNA if the ribosome pauses then specific bands will be improved in the trailing edge of the ribosome.[8]
Some of the elongation inhibitors, such as: cycloheximide (in eukaryotes) or chloramphenicol, cause the ribosomes to pause and to accumulate in the start codons. Elongation Factor P regulates the ribosomal pause at polyproline in bacteria, and when there is no EFP the density of ribosomes decreases from the polyproline motifs. If there are multiple ribosome pauses, then the EFP won't resolve it.[9]
Resolution and effects on gene expression
editSome forms of ribosomal pause are reversible without needing to discard the translated peptide and mRNA. This sort, usually described as a slowdown, is usually caused by polyproline stretches (resolved by EFP or eIF5A) and uncharged tRNA.[4] Slowdowns are important for the cell to control how much protein is produced;[10] it also aids co-translational folding of the nascent polypeptide on the ribosome, and delays protein translation while its encoding mRNA; this can trigger ribosomal frameshifting.[11]
More severe "stalls" can be caused an actual lack of tRNA or by the mRNA terminating without a stop codon.[4] In this case, ribosomal quality control (RQC) performs crisis rescue by translational abandonment. This releases the ribosome from the mRNA. The incomplete polypeptide is targeted for destruction; in eukaryotes, mRNA no-go decay is also triggered.[11]
It is difficult for RQC machinery to differentiate between a slowdown and a stall. It is possible for a mRNA sequence that normally produces a protein slowly to produce nothing instead due to intervention by RQC under different conditions.[12]
Rescue mechanisms
editIn bacteria, three rescue mechanisms are known.
- The main, universal system involves transfer-messenger RNA (tmRNA) and SmpB. The tRNA first binds to the ribosome like a tRNA, then with SmpB's help shifts into the mRNA position to translate a short peptide ending on a normal stop codon.[4]
- Alternative ribosome-rescue factor A (ArfA) is an alternative system in E. coli. It recruits RF2.[4]
- Alternative ribosome-rescue factor B (ArfB) is another alternative from E. coli. It works like a GGQ-release factor itself, releasing the peptide from tRNA.[13] At the same time, it fits into the mRNA tunnel to remove the mRNA.[14]
Advantage of the ribosomal pause
editWhen the ribosome movement on the mRNA is not linear, the ribosome gets paused at different regions without a precise reason. The ribosome pause position will help to identify the mRNA sequence features, structure, and the transacting factor that modulates this process.[15] The advantage of ribosomal pause sites that are located at protein domain boundaries are aiding the folding of a protein.[1] There are times when the ribosomal pause does not cause an advantage and it needs to be restricted. In translation, elF5A inhibits ribosomal pausing for translation to function better. Ribosomal pausing can cause more non-canonical start codons without elF5A in eukaryotic cells. When there is a lack of elF5A in the eukaryotic cell, it can cause an increase in ribosomal pausing.[16] The ribosomal pausing process can also be used by amino acids to control translation.[10]
The location of the ribosome pause event in vitro
editIt is known that ribosomes pause at distinct sites, but the reasons for these pauses are mostly unknown. Also, the ribosome pauses if the pseudoknot is disrupted. 10% of the ribosome pauses at the pseudoknot and 4% of the ribosomes are terminated. Before the ribosome is obstructed it passes the pseudoknot.[17] An assay was put together by a group from the University of California in an effort to show a model of mRNA. The translation was monitored in two in vitro systems. It was found that translating ribosomes aren't uniformly distributed along an mRNA.[8] Protein folding in vivo is also important and is related to protein synthesis. For finding the location of the ribosomal pause in vivo, the methods that have been used to find the ribosomal pause in vitro can be changed to find these specific locations in vivo.[6]
Ribosome profiling
editRibosome profiling is a method that can reveal pausing sites through sequencing the ribosome protected fragments (RPFs or footprints) to map ribosome occupancy on the mRNA. Ribosome profiling has the ability to reveal the ribosome pause sites in the whole transcriptome. When the kinetics layer is added,[18] it discloses the time of the pause, and the translation takes place.[9] Ribosome profiling is however still in early stages and has biases that need to be explored further.[19] Ribosome profiling allows for translation to be measured more accurately and precisely. During this process, translation needs to be stopped in order for ribosome profiling to be performed. This may cause a problem with ribosome profiling because the methods that are used to stop translation in an experiment can impact the outcome, which causes incorrect results. Ribosome profiling is useful for getting specific information on translation and the process of protein synthesis.[20]
See also
editReferences
edit- ^ a b Gawroński P, Jensen PE, Karpiński S, Leister D, Scharff LB (March 2018). "Pausing of Chloroplast Ribosomes Is Induced by Multiple Features and Is Linked to the Assembly of Photosynthetic Complexes". Plant Physiology. 176 (3): 2557–2569. doi:10.1104/pp.17.01564. PMC 5841727. PMID 29298822.
- ^ a b Li GW, Oh E, Weissman JS (March 2012). "The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria". Nature. 484 (7395): 538–41. Bibcode:2012Natur.484..538L. doi:10.1038/nature10965. PMC 3338875. PMID 22456704.
- ^ Lopinski JD, Dinman JD, Bruenn JA (February 2000). "Kinetics of ribosomal pausing during programmed -1 translational frameshifting". Molecular and Cellular Biology. 20 (4): 1095–103. doi:10.1128/MCB.20.4.1095-1103.2000. PMC 85227. PMID 10648594.
- ^ a b c d e f Buskirk, Allen R.; Green, Rachel (19 March 2017). "Ribosome pausing, arrest and rescue in bacteria and eukaryotes". Philosophical Transactions of the Royal Society B: Biological Sciences. 372 (1716): 20160183. doi:10.1098/rstb.2016.0183. PMC 5311927. PMID 28138069.
- ^ Kontos H, Napthine S, Brierley I (December 2001). "Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency". Molecular and Cellular Biology. 21 (24): 8657–70. doi:10.1128/MCB.21.24.8657-8670.2001. PMC 100026. PMID 11713298.
- ^ a b Jha SS, Komar AA (July 2012). "Isolation of ribosome bound nascent polypeptides in vitro to identify translational pause sites along mRNA". Journal of Visualized Experiments (65). doi:10.3791/4026. PMC 3471273. PMID 22806127.
- ^ Kim JK, Hollingsworth MJ (October 1992). "Localization of in vivo ribosome pause sites". Analytical Biochemistry. 206 (1): 183–8. doi:10.1016/s0003-2697(05)80031-4. PMID 1456432.
- ^ a b Wolin SL, Walter P (November 1988). "Ribosome pausing and stacking during translation of a eukaryotic mRNA". The EMBO Journal. 7 (11): 3559–69. doi:10.1002/j.1460-2075.1988.tb03233.x. PMC 454858. PMID 2850168.
- ^ a b Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS (February 2012). "High-resolution view of the yeast meiotic program revealed by ribosome profiling". Science. 335 (6068): 552–7. Bibcode:2012Sci...335..552B. doi:10.1126/science.1215110. PMC 3414261. PMID 22194413.
- ^ a b Darnell AM, Subramaniam AR, O'Shea EK (July 2018). "Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells". Molecular Cell. 71 (2): 229–243.e11. doi:10.1016/j.molcel.2018.06.041. PMC 6516488. PMID 30029003.
- ^ a b Buchan JR, Stansfield I (September 2007). "Halting a cellular production line: responses to ribosomal pausing during translation". Biology of the Cell. 99 (9): 475–87. doi:10.1042/BC20070037. PMID 17696878.
- ^ Collart MA, Weiss B (February 2020). "Ribosome pausing, a dangerous necessity for co-translational events". Nucleic Acids Research. 48 (3): 1043–1055. doi:10.1093/nar/gkz763. PMC 7026645. PMID 31598688.
- ^ Chan, KH; Petrychenko, V; Mueller, C; Maracci, C; Holtkamp, W; Wilson, DN; Fischer, N; Rodnina, MV (14 August 2020). "Mechanism of ribosome rescue by alternative ribosome-rescue factor B." Nature Communications. 11 (1): 4106. Bibcode:2020NatCo..11.4106C. doi:10.1038/s41467-020-17853-7. PMC 7427801. PMID 32796827.
- ^ Carbone, Christine E.; Demo, Gabriel; Madireddy, Rohini; Svidritskiy, Egor; Korostelev, Andrei A. (December 2020). "ArfB can displace mRNA to rescue stalled ribosomes". Nature Communications. 11 (1): 5552. Bibcode:2020NatCo..11.5552C. doi:10.1038/s41467-020-19370-z. PMC 7641280. PMID 33144582.
- ^ Wolin SL, Walter P (November 1988). "Ribosome pausing and stacking during translation of a eukaryotic mRNA". The EMBO Journal. 7 (11): 3559–69. doi:10.1002/j.1460-2075.1988.tb03233.x. PMC 454858. PMID 2850168.
- ^ Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, Mendell JT (December 2019). "Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection". Cell Reports. 29 (10): 3134–3146.e6. doi:10.1016/j.celrep.2019.10.129. PMC 6917043. PMID 31801078.
- ^ Somogyi P, Jenner AJ, Brierley I, Inglis SC (November 1993). "Ribosomal pausing during translation of an RNA pseudoknot". Molecular and Cellular Biology. 13 (11): 6931–40. doi:10.1128/mcb.13.11.6931. PMC 364755. PMID 8413285.
- ^ Lopinski, John D.; Dinman, Jonathan D.; Bruenn, Jeremy A. (2000). "Kinetics of Ribosomal Pausing during Programmed −1 Translational Frameshifting". Molecular and Cellular Biology. 20 (4): 1095–1103. doi:10.1128/mcb.20.4.1095-1103.2000. PMC 85227. PMID 10648594.
- ^ Buskirk AR, Green R (March 2017). "Ribosome pausing, arrest and rescue in bacteria and eukaryotes". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1716): 20160183. doi:10.1098/rstb.2016.0183. PMC 5311927. PMID 28138069.
- ^ Brar GA, Weissman JS (November 2015). "Ribosome profiling reveals the what, when, where and how of protein synthesis". Nature Reviews. Molecular Cell Biology. 16 (11): 651–64. doi:10.1038/nrm4069. PMC 5522010. PMID 26465719.