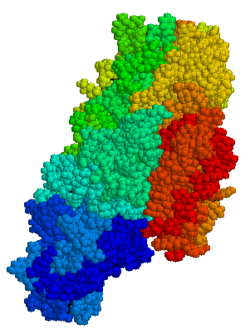
Tissue transglutaminase (abbreviated as tTG or TG2) is a 78-kDa, calcium-dependent enzyme (EC 2.3.2.13) of the protein-glutamine γ-glutamyltransferases family (or simply transglutaminase family).[5][6] Like other transglutaminases, it crosslinks proteins between an ε-amino group of a lysine residue and a γ-carboxamide group of glutamine residue, creating an inter- or intramolecular bond that is highly resistant to proteolysis (protein degradation). Aside from its crosslinking function, tTG catalyzes other types of reactions including deamidation, GTP-binding/hydrolyzing, and isopeptidase activities.[7] Unlike other members of the transglutaminase family, tTG can be found both in the intracellular and the extracellular spaces of various types of tissues and is found in many different organs including the heart, the liver, and the small intestine. Intracellular tTG is abundant in the cytosol but smaller amounts can also be found in the nucleus and the mitochondria.[6] Intracellular tTG is thought to play an important role in apoptosis.[8] In the extracellular space, tTG binds to proteins of the extracellular matrix (ECM),[9] binding particularly tightly to fibronectin.[10] Extracellular tTG has been linked to cell adhesion, ECM stabilization, wound healing, receptor signaling, cellular proliferation, and cellular motility.[6]
| Protein-glutamine gamma-glutamyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.3.2.13 | ||||||||
| CAS no. | 80146-85-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
tTG is the autoantigen in celiac disease, a lifelong illness in which the consumption of dietary gluten causes a pathological immune response resulting in the inflammation of the small intestine and subsequent villous atrophy.[11][12][13] It has also been implicated in the pathophysiology of many other diseases, including such as many different cancers and neurogenerative diseases.[14]
Structure
editGene
editThe human tTG gene is located on the 20th chromosome (20q11.2-q12).
Protein
editTG2 is a multifunctional enzyme that belongs to transglutaminases which catalyze the crosslinking of proteins by epsilon-(gamma-glutamyl)lysine isopeptide bonds.[15] Similarly to other transglutaminases, tTG consists of a GTP/ GDP binding site, a catalytic domain, two beta barrel and a beta-sandwich.[16] Crystal structures of TG2 with bound GDP, GTP, or ATP have demonstrated that these forms of TG2 adopt a "closed" conformation, whereas TG2 with the active site occupied by an inhibitory gluten peptide mimic or other similar inhibitors adopts an "open" conformation.[17][18][19] In the open conformation the four domains of TG2 are arranged in an extended configuration, allowing for catalytic activity, whereas in the closed conformation the two C-terminal domains are folded in on the catalytic core domain which includes the residue Cys-277.[20] The N-terminal domain only shows minor structural changes between the two different conformations.[21]
Mechanism
editThe catalytic mechanism for crosslinking in human tTG involves the thiol group from a Cys residue in the active site of tTG.[6] The thiol group attacks the carboxamide of a glutamine residue on the surface of a protein or peptide substrate, releasing ammonia, and producing a thioester intermediate. The thioester intermediate can then be attacked by the surface amine of a second substrate (typically from a lysine residue). The end product of the reaction is a stable isopeptide bond between the two substrates (i.e. crosslinking). Alternatively, the thioester intermediate can be hydrolyzed, resulting in the net conversion of the glutamine residue to glutamic acid (i.e. deamidation).[6] The deamidation of glutamine residues catalyzed by tTG is thought to be linked to the pathological immune response to gluten in celiac disease.[12] A schematic for the crosslinking and the deamidation reactions is provided in Figure 1.
Regulation
editThe expression of tTG is regulated at the transcriptional level depending on complex signal cascades. Once synthesized, most of the protein is found in the cytoplasm, plasma membrane and ECM, but a small fraction is translocated to the nucleus, where it participates in the control of its own expression through the regulation of transcription factors.[22]
Crosslinking activity by tTG requires the binding of Ca2+ ions.[23] Multiple Ca2+ can bind to a single tTG molecule.[6] Specifically, tTG binds up to 6 calcium ions at 5 different binding sites. Mutations to these binding sites causing lower calcium affinity, decrease the enzyme's transglutaminase activity.[14] In contrast, the binding of one molecule of GTP or GDP inhibits the crosslinking activity of the enzyme.[23] Therefore, intracellular tTG is mostly inactive due to the relatively high concentration of GTP/GDP and the low levels of calcium inside the cell.[6][12] Although extracellular tTG is expected to be active due to the low concentration of guanine nucleotides and the high levels of calcium in the extracellular space, evidence has shown that extracellular tTG is mostly inactive.[6][12][23] Recent studies suggest that extracellular tTG is kept inactive by the formation of a disulfide bond between two vicinal cysteine residues, namely Cys 370 and Cys 371.[24] When this disulfide bond forms, the enzyme remains in an open confirmation but becomes catalytically inactive.[24] The, oxidation/reduction of the disulfide bond serves as a third allosteric regulatory mechanism (along with GTP/GDP and Ca2+) for the activation of tTG.[12] Thioredoxin-1 has been shown to activate extracellular tTG by reducing the disulfide bond.[23] Another disuplhide bond can form in tTG, between the residues Cys-230 and Cys-370. While this bond does not exist in the enzyme's native state, it appears when the enzyme is inactivated via oxidation.[20] The presence of calcium protects against the formation of both disulfide bonds, thus making the enzyme more resistant to oxidation.[20]
Recent studies have suggested that interferon-γ may serve as an activator of extracellular tTG in the small intestine; these studies have a direct implication to the pathogenesis of celiac disease.[12] Activation of tTG has been shown to be accompanied by large conformational changes, switching from a compact (inactive) to an extended (active) conformation. (see Figure 3)[23][25][26]
In the extracellular matrix, TG2 is "turned off", due primarily to the oxidizing activity of endoplasmic reticulum protein 57 (ERp57).[24] Thus, tTG is allosterically regulated by two separate proteins, Erp57 and TRX-1.[24] (See Figure 4).
Function
edittTG is expressed ubiquitously and is present in various cellular compartments, such as the cytosol, the nucleus, and the plasma membrane.[14] It requires calcium as a cofactor for transamidation activity. Transcription is increased by retinoic acid. Among its many supposed functions, it appears to play a role in wound healing, apoptosis, and extracellular matrix development[11] as well as differentiation and cell adhesion.[14] It has been noted that tTG may have very different activity in different cell types. For example, in neurons, tTG supports the survival of cells subjected to injury whereas in astrocytes knocking out the gene expression for tTG is beneficial to cell survival.[27]
tTG is thought to be involved in the regulation of the cytoskeleton by crosslinking various cytoskeletal proteins including myosin, actin, and spectrin.[28] Evidence shows that intracellular tTG crosslinks itself to myosin. It is also believed that tTG may stabilize the structure of the dying cells during apoptosis by polymerizing the components of the cytoskeleton, therefore preventing the leakage of the cellular contents into the extracellular space.[7]
tTG also has GTPase activity:[5] In the presence of GTP, it suggested to function as a G protein participating in signaling processes.[29] Besides its transglutaminase activity, tTG is proposed to also act as kinase,[30] and protein disulfide isomerase,[31] and deamidase.[32] This latter activity is important in the deamidation of gliadin peptides, thus playing important role in the pathology of coeliac disease.
tTG also presents PDI (Protein Disulfide Isomerase) activity.[33][34] Based on its PDI activity, tTG plays an important role in the regulation of proteostasis, by catalyzing the trimerization of HSF1 (Heat Shock Factor 1) and thus the body's response to heat shock. In the absence of tTG, the response to heat shock is impaired since the necessary trimer is not formed.[34]
Clinical significance
edittTG is the most comprehensively studied transglutaminase and has been associated with many diseases. However, none of these diseases are related to an enzyme deficiency. Indeed, thus far no disease has been attributed to the lack of tTG activity and this has been attested through the study of tTG knockout mice.[35]
Celiac Disease
edittTG is best known for its link with celiac disease.[13] It was first associated with celiac disease in 1997 when the enzyme was found to be the antigen recognized by the antibodies specific to celiac.[35] Anti-transglutaminase antibodies result in a form of gluten sensitivity in which a cellular response to Triticeae glutens that are crosslinked to tTG are able to stimulate transglutaminase specific B-cell responses that eventually result in the production of anti-transglutaminase antibodies IgA and IgG.[36][37] tTG specifically deamidates the glutamine residues creating epitopes that increase the binding affinity of the gluten peptide to the antigen presenting T cells, initiating an adaptive immune response.[35]
Cancer
editRecent studies suggest that tTG also plays a role in inflammation and tumor biology.[11] tTG expression is elevated in multiple cancer cell types and is implicated in drug resistance and metastasis due to its ability to promote mesenchymal transition and stem cell like properties. In its GTP bound form, tTG contributes to cancer cell survival and appears to be a cancer driver. tTG is upregulated in cancer cells and tissues in many cancer types, including leukemia, breast cancer, prostate cancer, pancreatic cancer and cervical cancer. Higher tTG expression also correlates with higher instances of metastasis, chemotherapy resistance, lower survival rates and generally poor prognosis. Cancer cells can be killed by increasing calcium levels through the activation of tTG transamidation activity. Preclinical trials have showed promise in using tTG inhibitors as anti-cancer therapeutic agents.[38] However, other studies [33] have noted that tTG transamidation activity could be linked to the inhibition of tumor cell invasiveness.
Other Diseases
edittTG is believed to contribute to several neurodegenerative disorders including Alzheimer, Parkinson and Huntington diseases by affecting transcription, differentiation and migration and adhesion .[39][40] Such neurological diseases are characterized in part by the abnormal aggregation of proteins due to the increased activity of protein crosslinking in the affected brain.[41] Additionally, specific proteins associated with these disorders have been found to be in vivo and in vitro substrates of tTG.[7] Although tTG is up regulated in the areas of the brain affected by Huntington's disease, a recent study showed that increasing levels of tTG do not affect the onset and/or progression of the disease in mice.[42] Recent studies show that tTG may not be involved in AD as studies show it is associated with erythrocyte lysis and is a consequence of the disease rather than a cause.
tTG has also been linked to the pathogenesis of fibrosis in various organs including the lung and the kidney. Specifically, in kidney fibrosis, tTG contributes to the stabilization and accumulation of the ECM affecting TGF beta activity.[16]
Diagnostic
editSerology for anti-tTG antibodies has superseded older serological tests (anti-endomysium, anti-gliadin, and anti-reticulin) and has a strong sensitivity (99%) and specificity (>90%) for identifying celiac disease. Modern anti-tTG assays rely on a human recombinant protein as an antigen.[43]
Therapeutic
editIt's still experimental to use tTG as a form of surgical glue. It is also being studied as an attenuator of metastasis in certain tumors.[11] tTG shows promise as a potential therapeutic target to treat cardiac fibrosis, through the activity of a highly selective tTG inhibitor.[44] tTG inhibitors have also been shown to inhibit the formation of toxic inclusions related to neurodegenerative diseases.[45] This indicates that tTG inhibitors could also serve as a tool to mitigate the progression of tTG brain related diseases.[45]
Interactions
editTG2 participates in both enzymatic and non-enzymatic interactions. Enzymatic interactions are formed between TG2 and its substrate proteins containing the glutamine donor and lysine donor groups in the presence of calcium. Substrates of TG2 are known to affect TG2 activity, which enables it to subsequently execute diverse biological functions in the cell. However, the importance of non-enzymatic interactions in regulating TG2 activities is yet to be revealed. Recent studies indicate that non-enzymatic interactions play physiological roles and enable diverse TG2 functions in a context-specific manner.[46]
| Mouse Mutant Alleles for Tgm2 | |
|---|---|
| Marker Symbol for Mouse Gene. This symbol is assigned to the genomic locus by the MGI | Tgm2 |
| Mutant Mouse Embryonic Stem Cell Clones. These are the known targeted mutations for this gene in a mouse. | Tgm2tm1a(KOMP)Wtsi |
| Example structure of targeted conditional mutant allele for this gene | |
| These Mutant ES Cells can be studied directly or used to generate mice with this gene knocked out. Study of these mice can shed light on the function of Tgm2: see Knockout mouse | |
Erp57
editEndoplasmic reticulum protein 57 (Erp57), is a chaperone molecule involved in loading peptide onto MHC class I molecules in the endoplasmic reticulum.
Transglutaminase 2 (TG2) is a ubiquitously expressed (intracellular as well as extracellular) protein, with multiple modes of Post-translational regulation, including an allosteric disulfide bond between Cys-370-Cys-371 that renders the enzyme inactive in the extracellular matrix.[24]
Endoplasmic reticulum (ER)-resident protein 57 (ERp57), a protein in the ER that promotes folding of nascent proteins and is also present in the extracellular environment, has the cellular and biochemical characteristics for inactivating TG2. We found that ERp57 colocalizes with extracellular TG2 in cultured human umbilical vein endothelial cells (HUVECs). ERp57 oxidized TG2 with a rate constant that was 400-2000-fold higher than those of the aforementioned small molecule oxidants. Moreover, its specificity for TG2 was also markedly higher than those of other secreted redox proteins, including protein disulfide isomerase (PDI), ERp72, TRX, and quiescin sulfhydryl oxidase 1 (QSOX1).
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000198959 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000037820 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Király R, Demény M, Fésüs L (December 2011). "Protein transamidation by transglutaminase 2 in cells: a disputed Ca2+-dependent action of a multifunctional protein". The FEBS Journal. 278 (24): 4717–39. doi:10.1111/j.1742-4658.2011.08345.x. PMID 21902809. S2CID 19217277.
- ^ a b c d e f g h Klöck C, Diraimondo TR, Khosla C (July 2012). "Role of transglutaminase 2 in celiac disease pathogenesis". Seminars in Immunopathology. 34 (4): 513–22. doi:10.1007/s00281-012-0305-0. PMC 3712867. PMID 22437759.
- ^ a b c Facchiano F, Facchiano A, Facchiano AM (May 2006). "The role of transglutaminase-2 and its substrates in human diseases". Frontiers in Bioscience. 11: 1758–73. doi:10.2741/1921. PMID 16368554.
- ^ McConkey DJ, Orrenius S (October 1997). "The role of calcium in the regulation of apoptosis". Biochemical and Biophysical Research Communications. 239 (2): 357–66. CiteSeerX 10.1.1.483.2738. doi:10.1006/bbrc.1997.7409. PMID 9344835. S2CID 11242870.
- ^ Lortat-Jacob H, Burhan I, Scarpellini A, Thomas A, Imberty A, Vivès RR, Johnson T, Gutierrez A, Verderio EA (May 2012). "Transglutaminase-2 interaction with heparin: identification of a heparin binding site that regulates cell adhesion to fibronectin-transglutaminase-2 matrix". The Journal of Biological Chemistry. 287 (22): 18005–17. doi:10.1074/jbc.M111.337089. PMC 3365763. PMID 22442151.
- ^ Akimov SS, Krylov D, Fleischman LF, Belkin AM (February 2000). "Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin". The Journal of Cell Biology. 148 (4): 825–38. doi:10.1083/jcb.148.4.825. PMC 2169362. PMID 10684262.
- ^ a b c d Griffin M, Casadio R, Bergamini CM (December 2002). "Transglutaminases: nature's biological glues". The Biochemical Journal. 368 (Pt 2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- ^ a b c d e f Diraimondo TR, Klöck C, Khosla C (April 2012). "Interferon-γ activates transglutaminase 2 via a phosphatidylinositol-3-kinase-dependent pathway: implications for celiac sprue therapy". The Journal of Pharmacology and Experimental Therapeutics. 341 (1): 104–14. doi:10.1124/jpet.111.187385. PMC 3310700. PMID 22228808.
- ^ a b Di Sabatino A, Vanoli A, Giuffrida P, Luinetti O, Solcia E, Corazza GR (August 2012). "The function of tissue transglutaminase in celiac disease". Autoimmunity Reviews. 11 (10): 746–53. doi:10.1016/j.autrev.2012.01.007. PMID 22326684.
- ^ a b c d Király R, Csosz E, Kurtán T, Antus S, Szigeti K, Simon-Vecsei Z, Korponay-Szabó IR, Keresztessy Z, Fésüs L (December 2009). "Functional significance of five noncanonical Ca2+-binding sites of human transglutaminase 2 characterized by site-directed mutagenesis". The FEBS Journal. 276 (23): 7083–96. doi:10.1111/j.1742-4658.2009.07420.x. PMID 19878304. S2CID 21883387.
- ^ "Entrez Gene: TGM2 transglutaminase 2".
- ^ a b Hitomi K, Kojima S, Fesus L (2015). Transglutaminases : multiple functional modifiers and targets for new drug discovery. Tokyo. ISBN 9784431558255. OCLC 937392418.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ Pinkas DM, Strop P, Brunger AT, Khosla C (December 2007). "Transglutaminase 2 undergoes a large conformational change upon activation". PLOS Biology. 5 (12): e327. doi:10.1371/journal.pbio.0050327. PMC 2140088. PMID 18092889.
- ^ Liu S, Cerione RA, Clardy J (March 2002). "Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity". Proceedings of the National Academy of Sciences of the United States of America. 99 (5): 2743–7. Bibcode:2002PNAS...99.2743L. doi:10.1073/pnas.042454899. PMC 122418. PMID 11867708.
- ^ Han BG, Cho JW, Cho YD, Jeong KC, Kim SY, Lee BI (August 2010). "Crystal structure of human transglutaminase 2 in complex with adenosine triphosphate". International Journal of Biological Macromolecules. 47 (2): 190–5. doi:10.1016/j.ijbiomac.2010.04.023. PMID 20450932.
- ^ a b c Stamnaes J, Pinkas DM, Fleckenstein B, Khosla C, Sollid LM (August 2010). "Redox regulation of transglutaminase 2 activity". The Journal of Biological Chemistry. 285 (33): 25402–9. doi:10.1074/jbc.M109.097162. PMC 2919103. PMID 20547769.
- ^ Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM (August 2015). "Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease". The Journal of Biological Chemistry. 290 (35): 21365–75. doi:10.1074/jbc.M115.669895. PMC 4571865. PMID 26160175.
- ^ Bianchi N, Beninati S, Bergamini CM (May 2018). "Spotlight on the transglutaminase 2 gene: a focus on genomic and transcriptional aspects" (PDF). The Biochemical Journal. 475 (9): 1643–1667. doi:10.1042/BCJ20170601. hdl:11392/2388638. PMID 29764956.
- ^ a b c d e Jin X, Stamnaes J, Klöck C, DiRaimondo TR, Sollid LM, Khosla C (October 2011). "Activation of extracellular transglutaminase 2 by thioredoxin". The Journal of Biological Chemistry. 286 (43): 37866–73. doi:10.1074/jbc.M111.287490. PMC 3199528. PMID 21908620.
- ^ a b c d e Yi MC, Melkonian AV, Ousey JA, Khosla C (February 2018). "Endoplasmic reticulum-resident protein 57 (ERp57) oxidatively inactivates human transglutaminase 2". The Journal of Biological Chemistry. 293 (8): 2640–2649. doi:10.1074/jbc.RA117.001382. PMC 5827427. PMID 29305423.
- ^ Pinkas DM, Strop P, Brunger AT, Khosla C (December 2007). "Transglutaminase 2 undergoes a large conformational change upon activation". PLOS Biology. 5 (12): e327. doi:10.1371/journal.pbio.0050327. PMC 2140088. PMID 18092889.
- ^ Colak G, Keillor JW, Johnson GV (January 2011). Polymenis M (ed.). "Cytosolic guanine nucledotide binding deficient form of transglutaminase 2 (R580a) potentiates cell death in oxygen glucose deprivation". PLOS ONE. 6 (1): e16665. Bibcode:2011PLoSO...616665C. doi:10.1371/journal.pone.0016665. PMC 3031627. PMID 21304968.
- ^ Quinn BR, Yunes-Medina L, Johnson GV (July 2018). "Transglutaminase 2: Friend or foe? The discordant role in neurons and astrocytes". Journal of Neuroscience Research. 96 (7): 1150–1158. doi:10.1002/jnr.24239. PMC 5980740. PMID 29570839.
- ^ Nurminskaya MV, Belkin AM (2012). Cellular functions of tissue transglutaminase. International Review of Cell and Molecular Biology. Vol. 294. pp. 1–97. doi:10.1016/B978-0-12-394305-7.00001-X. ISBN 9780123943057. PMC 3746560. PMID 22364871.
- ^ Fesus L, Piacentini M (October 2002). "Transglutaminase 2: an enigmatic enzyme with diverse functions". Trends in Biochemical Sciences. 27 (10): 534–9. doi:10.1016/S0968-0004(02)02182-5. PMID 12368090.
- ^ Mishra S, Murphy LJ (June 2004). "Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase". The Journal of Biological Chemistry. 279 (23): 23863–8. doi:10.1074/jbc.M311919200. PMID 15069073.
- ^ Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y (August 2003). "A novel function of tissue-type transglutaminase: protein disulphide isomerase". The Biochemical Journal. 373 (Pt 3): 793–803. doi:10.1042/BJ20021084. PMC 1223550. PMID 12737632.
- ^ Sakly W, Thomas V, Quash G, El Alaoui S (December 2006). "A role for tissue transglutaminase in alpha-gliadin peptide cytotoxicity". Clinical and Experimental Immunology. 146 (3): 550–8. doi:10.1111/j.1365-2249.2006.03236.x. PMC 1810403. PMID 17100777.
- ^ a b Tabolacci C, De Martino A, Mischiati C, Feriotto G, Beninati S (January 2019). "The Role of Tissue Transglutaminase in Cancer Cell Initiation, Survival and Progression". Medical Sciences. 7 (2): 19. doi:10.3390/medsci7020019. PMC 6409630. PMID 30691081.
- ^ a b Rossin F, Villella VR, D'Eletto M, Farrace MG, Esposito S, Ferrari E, Monzani R, Occhigrossi L, Pagliarini V, Sette C, Cozza G, Barlev NA, Falasca L, Fimia GM, Kroemer G, Raia V, Maiuri L, Piacentini M (July 2018). "TG2 regulates the heat-shock response by the post-translational modification of HSF1". EMBO Reports. 19 (7): e45067. doi:10.15252/embr.201745067. PMC 6030705. PMID 29752334.
- ^ a b c Lorand L, Iismaa SE (January 2019). "Transglutaminase diseases: from biochemistry to the bedside". FASEB Journal. 33 (1): 3–12. doi:10.1096/fj.201801544R. PMID 30593123. S2CID 58551851.
- ^ Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, Schuppan D (July 1997). "Identification of tissue transglutaminase as the autoantigen of celiac disease". Nature Medicine. 3 (7): 797–801. doi:10.1038/nm0797-797. PMID 9212111. S2CID 20033968.
- ^ Murray JA, Frey MR, Oliva-Hemker M (June 2018). "Celiac Disease". Gastroenterology. 154 (8): 2005–2008. doi:10.1053/j.gastro.2017.12.026. PMC 6203336. PMID 29550590.
- ^ Eckert, Richard L. (2019-01-29). "Transglutaminase 2 takes center stage as a cancer cell survival factor and therapy target: Transglutaminase in cancer". Molecular Carcinogenesis. 58 (6): 837–853. doi:10.1002/mc.22986. PMC 7754084. PMID 30693974. S2CID 59341070.
- ^ Wilhelmus MM, Verhaar R, Andringa G, Bol JG, Cras P, Shan L, Hoozemans JJ, Drukarch B (March 2011). "Presence of tissue transglutaminase in granular endoplasmic reticulum is characteristic of melanized neurons in Parkinson's disease brain". Brain Pathology. 21 (2): 130–9. doi:10.1111/j.1750-3639.2010.00429.x. PMC 8094245. PMID 20731657. S2CID 586174.
- ^ Ricotta M, Iannuzzi M, Vivo GD, Gentile V (May 2010). "Physio-pathological roles of transglutaminase-catalyzed reactions". World Journal of Biological Chemistry. 1 (5): 181–7. doi:10.4331/wjbc.v1.i5.181. PMC 3083958. PMID 21541002.
- ^ Martin A, Giuliano A, Collaro D, De Vivo G, Sedia C, Serretiello E, Gentile V (January 2013). "Possible involvement of transglutaminase-catalyzed reactions in the physiopathology of neurodegenerative diseases". Amino Acids. 44 (1): 111–8. doi:10.1007/s00726-011-1081-1. PMID 21938398. S2CID 16143202.
- ^ Kumar A, Kneynsberg A, Tucholski J, Perry G, van Groen T, Detloff PJ, Lesort M (September 2012). "Tissue transglutaminase overexpression does not modify the disease phenotype of the R6/2 mouse model of Huntington's disease". Experimental Neurology. 237 (1): 78–89. doi:10.1016/j.expneurol.2012.05.015. PMC 3418489. PMID 22698685.
- ^ Sblattero D, Berti I, Trevisiol C, Marzari R, Tommasini A, Bradbury A, Fasano A, Ventura A, Not T (May 2000). "Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease". The American Journal of Gastroenterology. 95 (5): 1253–7. doi:10.1111/j.1572-0241.2000.02018.x. PMID 10811336. S2CID 11018740.
- ^ Wang Z, Stuckey DJ, Murdoch CE, Camelliti P, Lip GY, Griffin M (April 2018). "Cardiac fibrosis can be attenuated by blocking the activity of transglutaminase 2 using a selective small-molecule inhibitor". Cell Death & Disease. 9 (6): 613. doi:10.1038/s41419-018-0573-2. PMC 5966415. PMID 29795262.
- ^ a b Min B, Chung KC (January 2018). "New insight into transglutaminase 2 and link to neurodegenerative diseases". BMB Reports. 51 (1): 5–13. doi:10.5483/BMBRep.2018.51.1.227. PMC 5796628. PMID 29187283.
- ^ Kanchan K, Fuxreiter M, Fésüs L (August 2015). "Physiological, pathological, and structural implications of non-enzymatic protein-protein interactions of the multifunctional human transglutaminase 2". Cellular and Molecular Life Sciences. 72 (16): 3009–35. doi:10.1007/s00018-015-1909-z. PMC 11113818. PMID 25943306. S2CID 14849506.
External links
edit- Endomysial antibodies Archived 2021-05-12 at the Wayback Machine
- A collection of substrates and interaction partners of TG2 is accessible in the TRANSDAB, an interactive transglutaminase substrate database.