EosFP is a photoactivatable green to red fluorescent protein. Its green fluorescence (516nm) switches to red (581nm) upon UV irradiation of ~390nm (violet/blue light) due to a photo-induced modification resulting from a break in the peptide backbone near the chromophore.[2] Eos was first discovered as a tetrameric protein in the stony coral Lobophyllia hemprichii[3]. Like other fluorescent proteins, Eos allows for applications such as the tracking of fusion proteins, multicolour labelling and tracking of cell movement[4]. Several variants of Eos have been engineered for use in specific study systems including mEos2, mEos4 and CaMPARI.
History
editEosFP was first discovered in 2005 during a large scale screen for PAFPs (photoactivatable fluorescent proteins) within the stony coral Lobophyllia hemprichii.[3] It has since been successfully cloned in Escherichia coli and fusion constructs have been developed for use in human cells.[3] Eos was named after the Greek goddess of dawn.[3]
Unlike the tetrameric fluorescent proteins derived from anthozoan coral, which can interfere with normal cellular function due to interactions between protein subunits, EosFP has been broken up into dimeric and monomeric variants through the introduction of of single point mutations[5]. These variants have been successful in the tracking of cellular components without disturbing function in the host cell and maintain the same photophysical properties as wild-type Eos.[6]
Since their discovery, monomeric Eos probes (mEos) have been shown to localize in the cytosol, plasma membrane, endosomes, prevacuolar vesicles, vacuoles, the endoplasmic reticulum, golgi bodies, peroxisomes, mitochondria, invaginations, filamentous actin and cortical microtubules. [7] mEos fusion proteins allow for differential colour labelling in single cells, or groups of cells in developing organs. They can also be used for the understanding of spatial/ temporal interactions between organelles and vesicles. The two fluorescent forms of mEosFP (green and red) are compatible with CFP, GFP, YFP and RFP for multicolour labelling.[7]
Function
editEosFP emits a strong green fluorescence (516nm) that changes irreversibly to red (581nm) when irradiated with UV-light of 390nm. This modification occurs due to a break in the peptide backbone next to the chromophore.[3] This mechanism allows for localized tagging of the protein and makes EosFP an appropriate tool for tracking protein movement within living cells.[8] Formation of the red chromophore involves cleaving the peptide backbone but includes almost no other changes in the protein structure.[9]
According to single-molecule fluorescence spectroscopy , EosFP is tetrameric, and exhibits strong Forster resonance coupling within individual fluorophores.[3] Like other fluorescent proteins, Eos can be used to report diverse signals in cells, tissues and organs without disturbing complex biological machinery. While the use of fluorescent proteins was once limited to the green fluorescent protein (GFP), in recent years many other fluorescent proteins have been cloned. Unlike GFPs, which are derived from the luminescent jellyfish Aequorea victoria, fluorescent proteins derived from anthozoa, including Eos, emit fluorescence in the red spectral range. The novel property of photoinduced green-to-red conversion in Eos is useful because it allows for localized tracking of proteins in living cells.[3]EosFP is unique because it has a large separation in the wavelengths it can emit which allows for easy identification of peak colours.[6] All green-to-red photoinducible fluorescent proteins, including Eos, contain a chromophoric unit derived from the tripeptide his-tyr-gly. This green-to-red conversion is completed by light rather than chemical oxidation such as in other FPs.[6]
Structure and Absorbance Properties
editPrimary structure
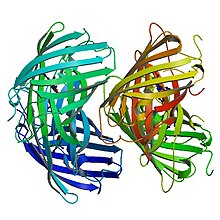
editEosFP consists of 226 amino acids. It has a molecular mass of 25.8 kDa and it's pl is 6.9. Eos has 84% identical residues to Kaede, a fluorescent protein that originated in a different scleractinian coral Trachyphyllia geoffroyi, but can also be irreversibly converted from a green to red emitting form using UV light.[9] Excluding residues Phe-61 and His-62, the chromophore environment and chromophore itself are unaffected by photochemical modification.[8] Wild-type EosFP has a tetrameric arrangement of subunits where each subunit has the same β-can structure as GFP. This structure includes an 11-stranded barrel and, down the central axis, the fluorophore-containing helix.
Structure of Green EosFP
editIn its anionic form, the green chromophore has an absorption maxima at 506nm and an emission maxima at 516nm. It is formed autocatalytically from amino acids His-62, Tyr-63 and Gly-64. Immediately surrounding the chromophore there is a cluster of charged or polar amino acids as well as structural water molecules. Above the plane of the chromophore, there is a network of hydrogen bond interactions between Glu-144, His-194, Glu-212 and Gln-38. Arg-66 and Arg-91 participate in hydrogen bonding with the carbonyl oxygen of green Eos's imidazolinone moiety. The His-62 side chain lies in an unpolar environment.[8] Conversion from the green to red form depends on the presence of a histidine in the first position of the tripeptide HYG that forms the chromophore. When this histidine residue is substituted with M, S, T or L, Eos only emits bright green light and no longer acts as a photoconvertible fluorescent protein.[3]
Structure of Red EosFP
editThe red chromophore, which is generated by cleavage of the peptide backbone, has an absorption maxima at 571nm and an emission maxima at 581nm, in its anionic form. The break in the peptide backbone that leads to this chromophore is between His-62 Nα and Cα.[6] The observed red fluorescence occurs due to an extension of the chromophore's π-conjugation where the His-62 imidazole ring connects to the imidazolinone. The hydrogen bond patterns of the red and green chromophores are almost identical.[8]
| Properties[10] | |
|---|---|
| Photoconversion wavelength | 390 nm |
| Green absorbance peak | 506 nm |
| Green emission peak | 516 nm |
| Red absorbance peak | 571 nm |
| Red emission peak | 581 nm |
| Green brightness* | 1.3X |
| Red brightness* | 0.7X |
| *Brightness values are relative to EGFP. | |
Photochemical conversion
editPhotochemical conversion occurs due to interactions between the chromophoric unit and residues in its vicinity. Glu-212 functions as a base that removes a proton from His-62 aiding in the cleavage of the His-62-Nα-Cα bond. Replacing Glu-212 with glutamine prevents photoconversion. At low pH, the yield of Eos involved in photoconversion is greatly increased as the fraction of molecules in the protonated form increases. The action spectrum for photoconversion is closely related to the action spectrum for Eos's protonated form. These observations suggest that the neutral form of the green chromophore, including a protonated Tyr-63 side chain, is the gateway structure for photoconversion. Proton ejection from the Tyr-63 phenyl side chain is an important event in the conversion mechanism where a proton is transferred from the His-62 imidazole, which is hydrogen-bonded to the Phe-61 carbonyl. The extra proton causes His-62 to donate a proton to the Phe-61 carbonyl forming a leaving group out of the peptide bond between His and Phe in the elimination reaction. The His-62 side chain is protonated during photoexcitation and assists the reaction by donating a proton to the Phe-61 carbonyl in the leaving group. After the backbone is cleaved, the hydrogen bond between His-62 and Phe-61 is reformed. When His-62 is replaced with other amino acids, EosFP loses its ability to photoconvert, providing evidence that His-62 is a necessary component of the photoconversion mechanism.[8] The internal charge distribution of the green chromophore is altered during photo excitation to assist in the elimination reaction. [6]
Spectroscopy
editBoth the fluorescence excitation and emission spectrums of wild-type EosFP are shifted ~65nm to the right upon excitation toward the red end of the spectrum. This spectral change is caused by an extension of the chromophore accompanied by a break in the peptide backbone between Phe-61 and His-62 in an irreversible mechanism[2]. The presence of a crisp isobestic point at 432nm also suggests an interconversion between two species. An absorption peak at 280nm is visible due to aromatic amino acids which transfer their excitation energy to the green chromophore. The quantum yield of the green-emitting form of Eos is 0.7[3]. In the red shifted species, there are pronounced vibronic sidebands separate from the main peak at 533nm and 629nm in the excitation spectrum and emission spectrum respectively. There is another peak in the red excitation spectrum at 502nm likely due to FRET excitation of the red fluorophore.[2] The quantum yield of the red-emitting form is 0.55.[3]
EosFPs variants show almost no difference in spectroscopic properties, therefore, it is likely that the structural modifications which arise from separation of interfaces have little to no effect on the structure of the fluorophore-binding site.[5]
Applications
editTracking of Fusion Proteins
editMany different fusion proteins have been created using EosFP and its engineered variants. These fusion proteins allow for the tracking of proteins within living cells while retaining complex biological functions like protein-protein interactions and protein-DNA interactions. Eos fusion constructs include those with recombination signal-binding protein (RBP) and cytokeratin.[3] Studies have shown that it is favourable to attach the protein of interest to the N-terminal side of the EosFP label[4]. These fusion constructs have been used to visualize nuclear translocation with androgen receptors, dynamics of the cytoskeleton with actin and vinculin and intranuclear protein movement with RBP.[4]
Multicolour Labelling
editSince EosFP can be used in fusion constructs while maintaining functionality of the protein of interest, it is a popular choice for multi-colour labelling studies.[4] In a dual-colour labelling experiment to map the stages of mitosis, HEK293 cells were first stably transfected with tubulin-binding protein cDNA fused to EGFP for visualization of the spindle apparatus. Then, transient transfection of recombination signal-binding protein (RBP) fused to d2EosFP was used to visualize the beginning of mitosis. Photoconversion was completed by fluorescent microscopy and highlighted the separation between two sets of chromosomes during anaphase, telophase and cytokinesis.[5]
Tracking of Cell Movement in Developmental Biology
editEosFP has been used to track cell movements during embryonic development of Xenopus laevis. At the two-cell/ early gastrula stage, capped mRNA coding for a dimeric EosFP (d2EosFP) was injected into cells and locally photoconverted using fluorescence microscopy[4]. These fluorescent embryos demonstrated the dynamics of cell movement during neurulation. EosFP was found in part of the notochord which shows the possibility of EosFP to be used in fate-mapping experiments.[5]
Engineered variants
editmEos4
editMany new monomeric versions of EosFP have been developed that offer advantages over wild type EosFP. Developed by a team at the Janelia Farm Research Campus at Howard Hughes Medical Institute, mEos4 has higher photostability and longer imaging abilities than EosFP. It is also highly resistant to chemical fixatives such as PFA, gluteraldehyde and OsO4 which are used to preserve samples. mEos4 is effective at higher temperatures than EosFP, phot-converts at an increased rate and has a higher emission amplitude in both green and red fluorescent states. Applications for the mEos4 protein include photoactivation localization microscopy (PALM), correlative light/ electron microscopy (CLEM), protein activity indication and activity integration (post-hoc imaging for protein activity over time).[11]
mEos2
editmEosFP is another monomeric Eos variant that folds effectively at 37 degrees celsius. Where tdEos (tandem dimer) cannot fuse to targets such as histones, tubulin, intermediate filaments and gap junctions, and mEos (monomeric) which can only be used successfully at 30 degrees celsius, mEos2 is an engineered variant that can fold effectively at 37 degrees celsius and successfully label targets intolerant to fusion from other fluorescent protein dimers . mEos2 shows almost identical spectral properties, brightness, pKa, photoconversion, contrast and maturation properties to WT Eos. The localization precision of mEos2 is twice as great as other monomeric fluorescent proteins.[11]
CaMPARI
editAlso at the Janelia Research Campus, a new fluorescent molecules known as CaMPARI (calcium-modulated photoactivatable ratiometric integrator) was developed using EosFP. The permanent green to red conversion signal was coupled with a calcium-sensitive protein, calmodulin, so that colour change in the fusion construct depended on the release of calcium accompanied by neural activity. CaMPARI is able to permanently mark neurons that are active at a any time. This visualization is possible across a wide amount of brain tissue as opposed to the limited view available with using a microscope. It also allows for the visualization of neural activity during complicated behaviours as the organism under study is allowed to move freely, rather than under a microscope. It also allows for the observation of neurons during specific behaviour periods. CaMPARI has, thus far, been used to label active neural circuits in mice, zebrafish and fruit flies.
Past Wiki Assignments
editWeek 5 - Possible Articles Ideas
Transcriptional Activator - Rated as a start class article of mid-importance in the wiki genetics project, this article has a decent amount of information but only one cited source. There is plenty of academic literature on activators and much that could be added to this article.
G-banding - Another start-class article, G-banding is used to produce karyotypes and identify chromosomal abnormalities. There seems to be a lot of literature on how G-banding can be used to diagnose chromosomal disorders and this information would be a useful addition to the article.
Mutational Meltdown - a popular topic in conservation genetics, mutational meltdown is discussed in many research articles regarding population genetics. This article also has only one reference and could be improved upon with additional credible information.
Chiasma - this article specifically needs the addition of research in chiasma interference. It appears as someone had previously tried to add information about research in this area but did not have enough information to make a meaningful addition and so their work was left on the talk page.
Week 4 Assignment - Added to the article (stub) 3-Iodotyrosine with citations.
Week 3 Assignment - Article evaluation for Gene Flow.
- Is everything in the article relevant to the article topic? Is there anything that distracted you?
Everything in the article is related to the topic of gene flow and well-organized. The article begins with an easy to understand introduction on the topic of gene flow including a definition, factors that affect the rate of gene flow between populations and examples of populations that can and cannot exhibit the effects of gene flow (and why or why not). There is also a diagram created by a wikipedia user that shows how gene flow works which would be helpful for individuals who came to the page with no background knowledge of gene flow.
- Is the article neutral? Are there any claims, or frames, that appear heavily biased toward a particular position?
All viewpoints in the article are neutral and factual. After reviewing the talk page, it does seem as though there were some opinionated statements originally included in the article but have since bee removed by editors.
- Check a few citations. Do the links work? Does the source support the claims in the article?
All of the links within the citations seem to work however one of the citations is missing the article title. The sources are mainly comprised of primary research articles within the field of population genetics and gene transfer. Other legitimate sources include textbooks. One of the sources seems to be from a science website that has little credibility and might need to be replaced by a more legitimate source.
- Is any information out of date? Is anything missing that could be added?
I personally found that the article covered all of the topics within gene flow that I was looking for. The article highlighted the concepts of how to measure gene flow, barriers of gene flow, how gene flow can occur between species and genetic pollution. The article also highlighted interesting examples of gene flow within natural populations. If anything, I would like to see more examples within the article (which only showcased about 6) as there are so many relevant examples of gene flow especially in current world issues such as in GMO crops . However, keeping the number of examples to a limited number does keep the article concise so perhaps if more examples are to be added they should start their own sub-page.
- Check out the Talk page of the article. What kinds of conversations, if any, are going on behind the scenes about how to represent this topic?
As previously mentioned, the talk page highlighted a few sentences that took an opinionated approach and were subsequently removed by other users and replaced with neutral viewpoints. One user on the talk-page stated that they found the introduction too wordy and took the liberty of editing it to be more concise. Another user pointed out a few wrong statements which they removed and stated they would aim to find evidence against them (in primary research) so that the page could be updated with more acurate information.
- How is the article rated? Is it a part of any WikiProjects?
This article is rated as a start class, high importance and is part of the WikiProject Genetics and Evolutionary Biology.
Looks great! Keep it up! AdamCF87 (talk) 15:29, 17 October 2017 (UTC)
- ^ Nienhaus, K.; Nienhaus, G. U.; Wiedenmann, J.; Nar, H. (2005). "Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP". Proc.Natl.Acad.Sci.Usa. 102: 9156–9159. doi:10.2210/pdb1zux/pdb.
- ^ a b c Ivanchenko, Sergey; Röcker, Carlheinz; Oswald, Franz; Wiedenmann, Jörg; Nienhaus, G. Ulrich (2005). "Targeted Green-Red Photoconversion of EosFP, a Fluorescent Marker Protein". Journal of Biological Physics. 31 (3–4): 249–259. doi:10.1007/s10867-005-0174-z. ISSN 0092-0606. PMC 3456337. PMID 23345897.
{{cite journal}}: CS1 maint: PMC format (link) - ^ a b c d e f g h i j k Wiedenmann, Jörg; Ivanchenko, Sergey; Oswald, Franz; Schmitt, Florian; Röcker, Carlheinz; Salih, Anya; Spindler, Klaus-Dieter; Nienhaus, G. Ulrich (2004-11-09). "EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion". Proceedings of the National Academy of Sciences of the United States of America. 101 (45): 15905–15910. doi:10.1073/pnas.0403668101. ISSN 0027-8424. PMID 15505211.
- ^ a b c d e Wiedenmann, Jörg; Nienhaus, G Ulrich. "Live-cell imaging with EosFP and other photoactivatable marker proteins of the GFP family". Expert Review of Proteomics. 3 (3): 361–374. doi:10.1586/14789450.3.3.361.
- ^ a b c d Nienhaus, G. Ulrich; Nienhaus, Karin; Hölzle, Angela; Ivanchenko, Sergey; Renzi, Fabiana; Oswald, Franz; Wolff, Michael; Schmitt, Florian; Röcker, Carlheinz. "Photoconvertible Fluorescent Protein EosFP: Biophysical Properties and Cell Biology Applications". Photochemistry and Photobiology. 82 (2). doi:10.1562/2005-05-19-ra-533.
- ^ a b c d e Mizuno, Hideaki; Mal, Tapas Kumar; Tong, Kit I.; Ando, Ryoko; Furuta, Toshiaki; Ikura, Mitsuhiko; Miyawaki, Atsushi (2003-10-01). "Photo-Induced Peptide Cleavage in the Green-to-Red Conversion of a Fluorescent Protein". Molecular Cell. 12 (4): 1051–1058. doi:10.1016/S1097-2765(03)00393-9.
- ^ a b Mathur, Jaideep; Radhamony, Resmi; Sinclair, Alison M.; Donoso, Ana; Dunn, Natalie; Roach, Elyse; Radford, Devon; Mohaghegh, P. S. Mohammad; Logan, David C. (2010-12-01). "mEosFP-Based Green-to-Red Photoconvertible Subcellular Probes for Plants". Plant Physiology. 154 (4): 1573–1587. doi:10.1104/pp.110.165431. ISSN 0032-0889. PMID 20940350.
- ^ a b c d e Nienhaus, Karin; Nienhaus, G. Ulrich; Wiedenmann, Jörg; Nar, Herbert (2005-06-28). "Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP". Proceedings of the National Academy of Sciences of the United States of America. 102 (26): 9156–9159. doi:10.1073/pnas.0501874102. ISSN 0027-8424. PMID 15964985.
- ^ a b Ando, Ryoko; Hama, Hiroshi; Yamamoto-Hino, Miki; Mizuno, Hideaki; Miyawaki, Atsushi (2002-10-01). "An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein". Proceedings of the National Academy of Sciences. 99 (20): 12651–12656. doi:10.1073/pnas.202320599. ISSN 0027-8424. PMID 12271129.
- ^ Lukyanov et al. 2005. Nature Reviews Molecular Cell Biology 6(11) pp 885-91.
- ^ a b "mEos2 & mEos4 | Janelia Research Campus". www.janelia.org. Retrieved 2017-11-28.
- ^ "New Fluorescent Protein Permanently Marks Neurons that Fire". HHMI.org. Retrieved 2017-12-01.