In molecular biology SprD (Small pathogenicity island RNA D) is a non-coding RNA expressed on pathogenicity islands in Staphylococcus aureus.[1] It was identified in silico along with a number of other sRNAs (SprA-G) through microarray analysis which were confirmed using a Northern blot.[2] SprD has been found to significantly contribute to causing disease in an animal model.[1]
| SprD | |
|---|---|
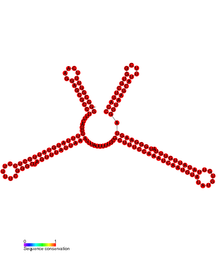 Conserved secondary structure of SprD. | |
| Identifiers | |
| Symbol | SprD |
| Rfam | RF01828 |
| Other data | |
| RNA type | Gene |
| Domain(s) | Staphylococcus aureus |
| PDB structures | PDBe |
Function
editSprD is located between genes scn and chp in the innate immune evasion cluster (IEC) of the S. aureus genome. Its placement within this region was the first indication of a virulence-factor regulatory function.[1]
SprD binds with sbi (Staphylococcus aureus binder of IgG)[3] mRNA which encodes an immune evasion protein. It occludes the Shine-Dalgarno sequence and the initiation codon of sbi, forming a sbi mRNA-SprD duplex repressing the translation of the mRNA.[1]
sbi protein interferes with the host's innate immune response by binding Factor H, Complement component 3 and IgG.[3][4]
See also
editReferences
edit- ^ a b c d Chabelskaya S, Gaillot O, Felden B (June 2010). "A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule". PLOS Pathog. 6 (6): e1000927. doi:10.1371/journal.ppat.1000927. PMC 2880579. PMID 20532214.
- ^ Pichon C, Felden B (October 2005). "Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains". Proc. Natl. Acad. Sci. U.S.A. 102 (40): 14249–14254. doi:10.1073/pnas.0503838102. PMC 1242290. PMID 16183745.
- ^ a b Haupt K, Reuter M, van den Elsen J, et al. (December 2008). "The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b". PLOS Pathog. 4 (12): e1000250. doi:10.1371/journal.ppat.1000250. PMC 2602735. PMID 19112495.
- ^ Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L (April 1998). "A second IgG-binding protein in Staphylococcus aureus". Microbiology. 144 (4): 985–991. doi:10.1099/00221287-144-4-985. PMID 9579072. Retrieved 2010-08-09.
Further reading
edit- Upadhyay A, Burman JD, Clark EA, et al. (August 2008). "Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi". J. Biol. Chem. 283 (32): 22113–22120. doi:10.1074/jbc.M802636200. PMC 2494919. PMID 18550524.
- Burman JD, Leung E, Atkins KL, et al. (June 2008). "Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus". J. Biol. Chem. 283 (25): 17579–17593. doi:10.1074/jbc.M800265200. PMC 2649420. PMID 18434316.
- Atkins KL, Burman JD, Chamberlain ES, et al. (March 2008). "S. aureus IgG-binding proteins SpA and Sbi: host specificity and mechanisms of immune complex formation". Mol. Immunol. 45 (6): 1600–1611. doi:10.1016/j.molimm.2007.10.021. PMID 18061675.
- Chabelskaya S, Bordeau V, Felden B (Apr 1, 2014). "Dual RNA regulatory control of a Staphylococcus aureus virulence factor". Nucleic Acids Research. 42 (8): 4847–4858. doi:10.1093/nar/gku119. PMC 4005698. PMID 24510101.